Team:TU Darmstadt/strategy
From 2013.igem.org
| Line 2: | Line 2: | ||
<style type="text/css"> | <style type="text/css"> | ||
| - | body | + | body |
{ | { | ||
margin:0; | margin:0; | ||
| Line 15: | Line 15: | ||
{ | { | ||
background-color: white; | background-color: white; | ||
| - | background-image: url("/wiki/images/ | + | background-image: url("/wiki/images/5/5c/Hintergrund_Grün(ohne_Header%2C_Logo_und_Schlagschatten).jpg"); |
background-attachment: scroll; | background-attachment: scroll; | ||
background-position: 50% 0; | background-position: 50% 0; | ||
| Line 26: | Line 26: | ||
{ | { | ||
background: white; | background: white; | ||
| - | background-image: url( | + | background-image: url("/wiki/images/2/2f/Header_%28mit_Logo%29.jpg"); |
margin: 0 auto; | margin: 0 auto; | ||
height:250px ; | height:250px ; | ||
| Line 35: | Line 35: | ||
background-repeat: no-repeat; | background-repeat: no-repeat; | ||
background-size: 100% 100% ; | background-size: 100% 100% ; | ||
| - | border-style: none; | + | border-style: none;;} |
| - | } | + | |
#p-logo { display:none;} | #p-logo { display:none;} | ||
| Line 108: | Line 107: | ||
position:absolute; | position:absolute; | ||
top:10px; | top:10px; | ||
| - | left: | + | left:500px; |
z-index: 5; | z-index: 5; | ||
} | } | ||
| + | |||
p, ul { | p, ul { | ||
| Line 116: | Line 116: | ||
margin: 0; | margin: 0; | ||
} | } | ||
| + | |||
</style> | </style> | ||
| + | |||
| + | <center> | ||
| + | <!-- central main menu --> | ||
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | |||
<!-- Taskbar --> | <!-- Taskbar --> | ||
| Line 126: | Line 134: | ||
<img alt="Problem" src="/wiki/images/6/66/Darmstadt_green_Problem.jpg" width="100" height="30"></a> | <img alt="Problem" src="/wiki/images/6/66/Darmstadt_green_Problem.jpg" width="100" height="30"></a> | ||
| - | |||
| - | |||
<a href="https://2013.igem.org/Team:TU_Darmstadt/result"> | <a href="https://2013.igem.org/Team:TU_Darmstadt/result"> | ||
| Line 140: | Line 146: | ||
<a href="https://2013.igem.org/Team:TU_Darmstadt/strategy"> | <a href="https://2013.igem.org/Team:TU_Darmstadt/strategy"> | ||
| - | <img alt="team" src="/wiki/images/a/ | + | <img alt="team" src="/wiki/images/a/a9/07._Strategy_(angewählt).jpg" width="100" height="30"></a> |
<a href="https://2013.igem.org/Team:TU_Darmstadt/humanpractice"> | <a href="https://2013.igem.org/Team:TU_Darmstadt/humanpractice"> | ||
| Line 150: | Line 156: | ||
<a href="https://2013.igem.org/Team:TU_Darmstadt/labbook"> | <a href="https://2013.igem.org/Team:TU_Darmstadt/labbook"> | ||
<img alt="team" src="/wiki/images/f/f3/Darmstadt_green_Labbook.jpg" width="90" height="30"></a> | <img alt="team" src="/wiki/images/f/f3/Darmstadt_green_Labbook.jpg" width="90" height="30"></a> | ||
| + | |||
| + | |||
| + | |||
</div> | </div> | ||
| + | |||
Revision as of 18:42, 4 October 2013
The Strategy
To easily detect biological toxins we developed a handy device, everybody is able to operate reliably.
By altering the sensorium of E. coli with the help of synthetic biology, the bacteria would release a defined beacon when sensing mycotoxins. This treatment relies on a change of the so called Tar receptors exterior binding site, to specifically react on mycotoxins [1].
By also changing the interior impulse, an explicit beacon can be measured using the interaction of two fluorescent colorants (FRET).
Samples can be given into a self-constructed capsule containing the modified bacteria.
The light-signal inside the capsule is measured using a low-cost and open-source constructed FRET analysing system, as well developed by our team.
To illustrate the data in a clear and usable way, the construction of an Android smartphone app is the completion of our project. The application should identify the capsule and apply an online test protocol in order to visualize and compare the data with former tests and share it with customers, leaving the consumer with the opportunity to check different producers on clean manufacturing and production. Consequently, food can be checked on mycotoxins in a cheap and reliable way.
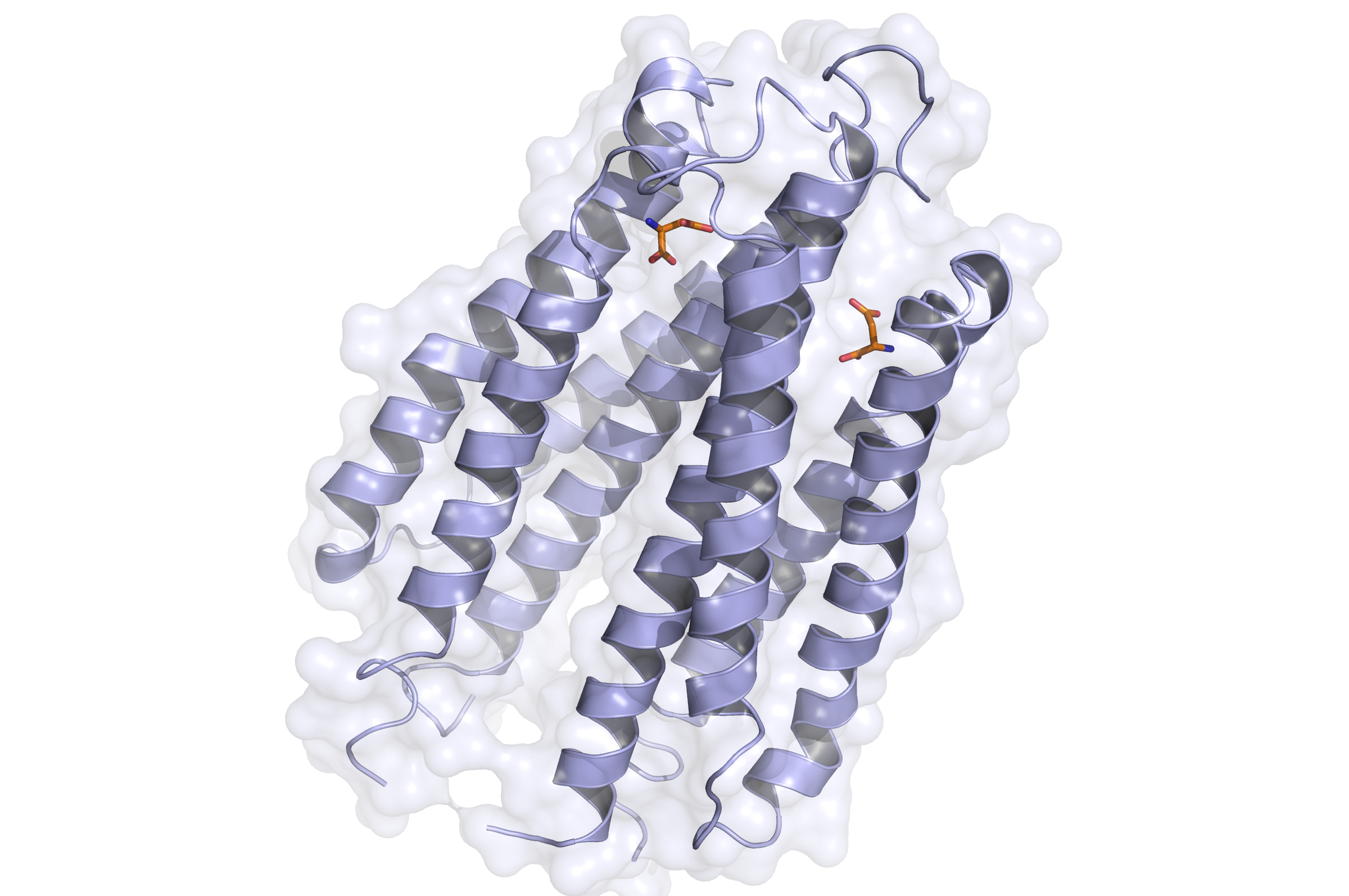
Tar Receptor and CheB
The Tar protein of Escherichia coli is a receptor protein, that forwards signals from outside into the cell. Its native task is to detect nutritive substances and to induce an active movement towards the source, a so called chemotaxis.
The receptor consists of two identical parts, called homologue monomers and is transmembrane.
The detection is taking place at the external part.
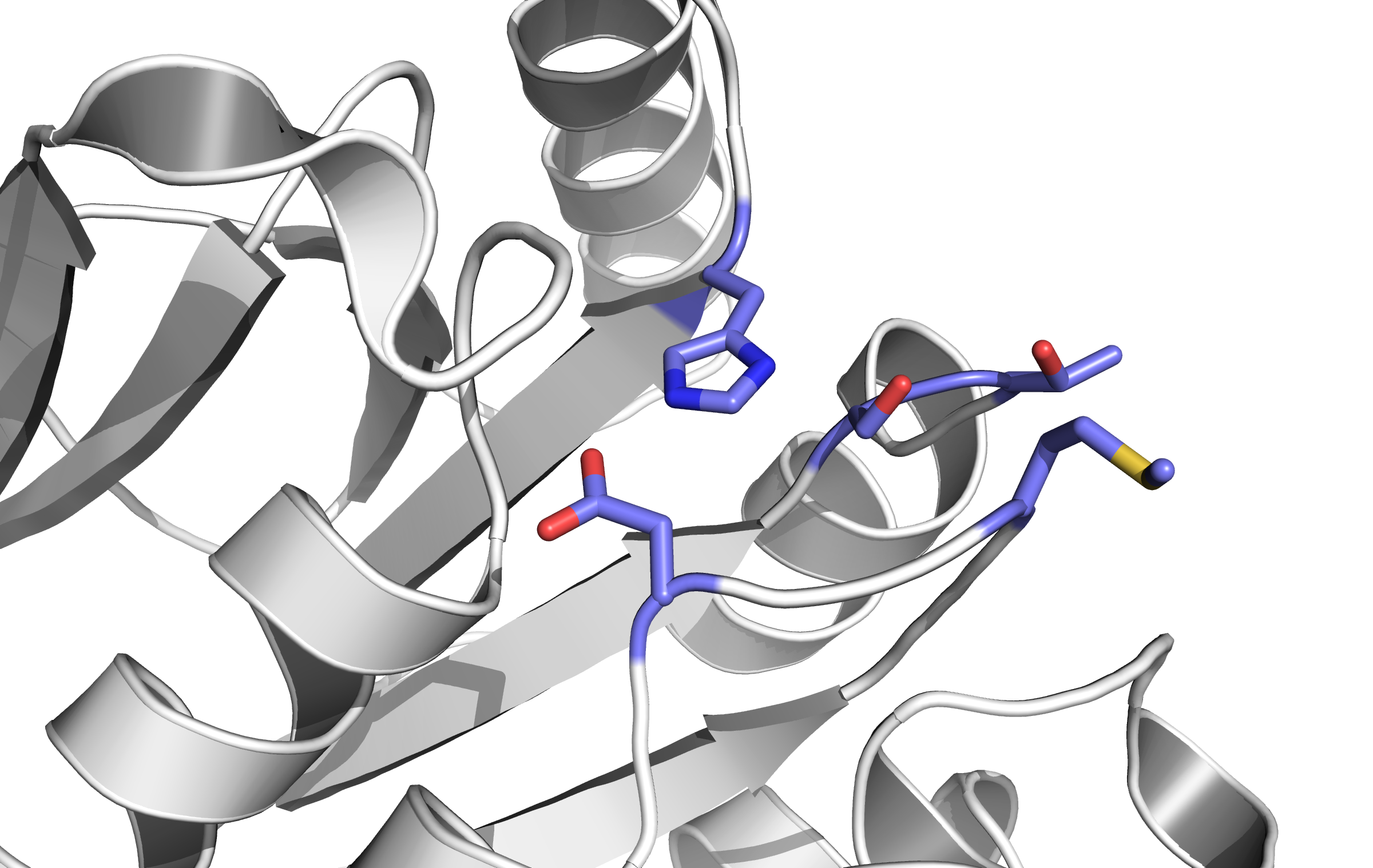
When detecting a substance, the receptor is switching form: a conformational change. This causes an alternation inside the cell - the receptor binds proteins transforming the signal into an inner cell cascade. Here, the first Protein binding to Tar is called CheB. These two build up the pairs needed for FRET signalling.
As the substance does not enter the cell, the Tar receptor is predestined for usage of toxin recognition.
Binding of the Tar receptor has to be absolutely specific to eliminate false positive signals. Therefore, in the external region of the receptor some amino acids have to be changed. A library, where the receptors are tested if the specification for a single toxin has improved, has to be created until a satisfactory version is found.
FRET
FRET (Förster or Flourescence energy transfer) is a technique to investigate protein interaction in living cells. We adapted this system to use it as a detection element for our selfmade FRET analyser. It works via two chromophores coupled to the prospective proteins. If we use light to excite the cells containing chromophore-coupled proteins, we can get two types of information: in our case the colour orange stands for a negative and red for a positive toxin result. This little trick analyses probes within seconds, hence detection works in real time. An important and often used FRET pair is LSSmOrange and mKate. Their excitation and emission spectra have a compatible overlap.
In detail: The resulting data gives information about the chromophores connected to the proteins and the distance and angle to each other. The principle relies on a radiation free transition: An excited donor transfers energy to the acceptor by dipole dipole interactions. Here, the given distance between the two molecules needs to be between 0,5 nm and 10 nm for an efficient FRET.
Using the dipole dipole interactions changes in physical structure, bindings and separations can be investigated contemporarily.
References
- Derr, P. B., 2006 Changing the Specificity of a Bacterial Chemoreceptor. Journal of Molecular Biology , 923–932
 "
"