Team:TU Darmstadt
From 2013.igem.org
| Line 157: | Line 157: | ||
<p text-aligne:left style="margin-left:10px; margin-right:10px"> | <p text-aligne:left style="margin-left:10px; margin-right:10px"> | ||
<br><br> | <br><br> | ||
| - | To easily detect biological toxins we developed a handy device, everybody is able to operate reliably. | + | To easily detect biological toxins we developed a <a href="https://2013.igem.org/Team:TU_Darmstadt/result/electrical_engineering">handy device</a>, everybody is able to operate reliably. |
</B><br><br><br> | </B><br><br><br> | ||
| Line 264: | Line 264: | ||
<br> | <br> | ||
<br> | <br> | ||
| - | Mycotoxins produced by mould fungus are present in our daily life and are harmful for humans as well as for animals. | + | <a href="https://2013.igem.org/Team:TU_Darmstadt/problem">Mycotoxins</a> produced by mould fungus are present in our daily life and are harmful for humans as well as for animals. |
The danger of contamination of grain and other natural products has fatal consequences for the economy and the supply of whole nations. | The danger of contamination of grain and other natural products has fatal consequences for the economy and the supply of whole nations. | ||
The mycotoxins which are permanently formed by the fungus, can be used as biomarkers to detect contamination. | The mycotoxins which are permanently formed by the fungus, can be used as biomarkers to detect contamination. | ||
| Line 270: | Line 270: | ||
To achieve this goal our team uses various methods from the fields of synthetic biology, | To achieve this goal our team uses various methods from the fields of synthetic biology, | ||
electrical engineering and information processing. The detection system relies on <i>E.coli</i> | electrical engineering and information processing. The detection system relies on <i>E.coli</i> | ||
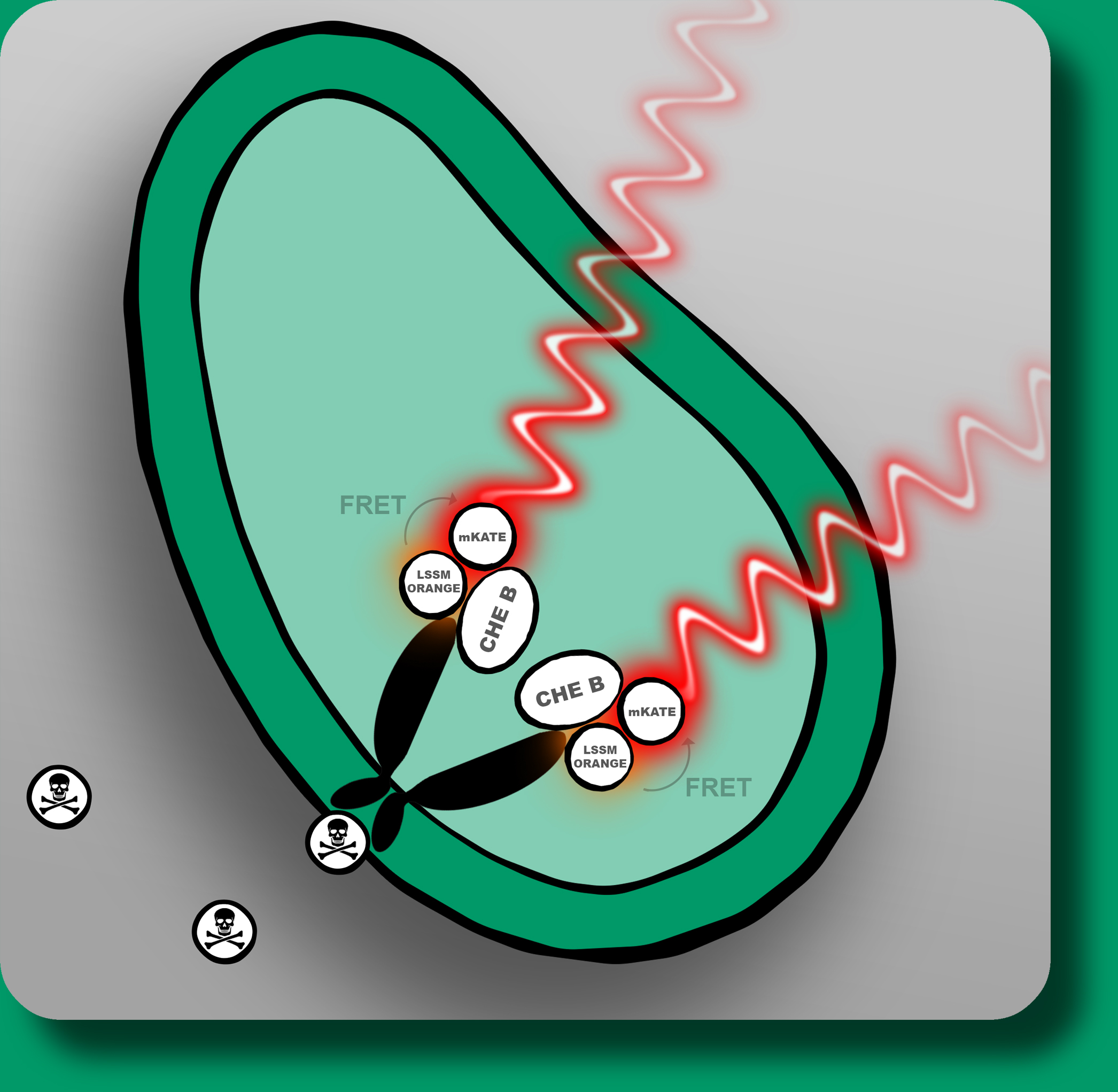
| - | with modified aspartate receptors (TAR) which interact with specific mycotoxins. | + | with modified aspartate receptors (<a href="https://2013.igem.org/Team:TU_Darmstadt/strategy>TAR</a>) which interact with specific mycotoxins. |
If these are present in the reviewed sample they will bind to the receptor and induce | If these are present in the reviewed sample they will bind to the receptor and induce | ||
a conformational change and thereby generate a measurable FRET-beacon by bringing | a conformational change and thereby generate a measurable FRET-beacon by bringing | ||
Revision as of 02:48, 5 October 2013

PROBLEM
It is estimated that about a quarter of the world’s food crops is affected by harmful mycotoxins produced by mould fungus.
FAO and WHO have developed three benchmarks of risk analysis: risk assessment, risk management and risk communication.
Our detection system covers those three benchmarks.
Click to read more.
STRATEGY
To easily detect biological toxins we developed a handy device, everybody is able to operate reliably.
Click to read more.

RESULT
In our results we show you all important informations about our work in the lab, with gorgeous graphs.
Click to read more.
Abstract
Mycotoxins produced by mould fungus are present in our daily life and are harmful for humans as well as for animals.
The danger of contamination of grain and other natural products has fatal consequences for the economy and the supply of whole nations.
The mycotoxins which are permanently formed by the fungus, can be used as biomarkers to detect contamination.
We want to develop a handy device which allows an easy and reliable detection of mycotoxins.
To achieve this goal our team uses various methods from the fields of synthetic biology,
electrical engineering and information processing. The detection system relies on E.coli
with modified aspartate receptors (
var _gaq = _gaq || [];
_gaq.push(['_setAccount', 'UA-43014839-1']);
_gaq.push(['_trackPageview']);
(function() {
var ga = document.createElement('script'); ga.type = 'text/javascript'; ga.async = true;
ga.src = ('https:' == document.location.protocol ? 'https://ssl' : 'http://www') + '.google-analytics.com/ga.js';
var s = document.getElementsByTagName('script')[0]; s.parentNode.insertBefore(ga, s);
}) ();
 "
"