Team:Wageningen UR/Notebook
From 2013.igem.org
(→notebook entries) |
(→notebook entries) |
||
| Line 355: | Line 355: | ||
</div> | </div> | ||
| - | div class="item human"> | + | <div class="item human"> |
<img src="http://sciencecafewageningen.nl/wp-content/uploads/2011/07/logoSCW.jpg"/> | <img src="http://sciencecafewageningen.nl/wp-content/uploads/2011/07/logoSCW.jpg"/> | ||
<div class="icon human"></div> | <div class="icon human"></div> | ||
Revision as of 12:13, 1 September 2013
- Safety introduction
- General safety
- Fungi-related safety
- Biosafety Regulation
- Safety Improvement Suggestions
- Safety of the Application
- Lablog
- Experimental protocols
notebook entries
Week 16
September (16.09 - 22.09)

Science Cafe Wageningen
16.09.2013
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Week 15
September (09.09 - 15.09)
Week 14
September (02.09 - 08.09)
Week 13
August (26.08 - 1.09)
Week 12
August (19.08 - 25.08)

iGEM Netherlands
24.08.2013
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.

Working on the Wiki
20.08.2013
The wiki is making good progress, everybody should be able to work with the system now. Objectives are to upload content, look for icons that can be used in the menu bar and to finish the team page

Transformations
19.08.2013
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.

G-blocks from IDT arrived today
19.08.2013
Today the G-blocks we ordered finally arrived! This is great news, because now we can start on the gibson assembly and transformations.

Lovastatin Pathway
19.08.2013
The lovastatin pathway has been added to the metabolic model of Aspergillus niger. Because the medium composition of the model is not yet properly defined, the maximum flux towards lovastatin can be achieved. This however is not realistic and therefore the next thing is to redefined the medium composition.
- B.D. Ames et al., 2011. Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis. PNAS, doi/10.1073/pnas.1113029109
Week 11
August (12.08 - 18.08)

Title
00.00.2013
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat.

Title
00.00.2013
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
Week 10
August (05.08 - 11.08)

Workshop by Paulien Poelarends
09.08.2013
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.

Protoplasts
12.08.2013
During this week we have prepared media and spores to make protoplasts. The protoplasts will be used for transformations. Until recently Novozym has been used to make protoplasts, this is not available anymore and some new protocols have to be used.

Lovastatin
06.08.2013
The restriction enzymes that we ordered last week have arrived. After a few attempts the sequences have been optimized and the G-blocks can be ordered. Also some Lovastatin has been ordered to perform some experiments with A. niger. Lovastatin may be fatal to cell growth of A. niger therefore experiments have to indicate whether A. niger is resistant to Lovastatin. Otherwise A. niger has to be transformed with the resistance gene from A. terreus.

Expanding the database
05.08.2013
In order to expand the database, more information on secondary metabolite backbone enzymes from Aspergilli needs to be added. Luckily there is a recent publication on secondary metabolites from A. nidulans, A. fumigatus, A. niger and A. oryzae.
- D.O. Inglis et al., 2013. Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiology, Vol. 13, p. 1-23.
Week 9
July (29.07 - 04.08)

Metabolic modeling
30.07.2013
The current metabolic model of A. niger needs to be expanded to include the lovastatin pathway as known from literature. First the model has to be checked: is it balanced? can we perform a FBA on the current model?
- M.R. Anderson et al., 2008. Metabolic model integration of the bibliome, genome, metabolome and reactome of Aspergillus niger. Molecular Systems Biology, Vol. 4, Article number 178; doi:10.1038/msb.2008.12

Shopping
30.07.2013
This week we ordered the restriction enzymes for the assembly strategy. Also other stuff should be ordered like pJET, lovastatin, A. niger & A. terrus strain, Gibson assembly kit...
Week 8
July (22.07 - 28.07)

Metabolic activity: GFP
24.07.2013
In a similar way that the DAPI stained cells give an indication of metabolic activity by showing an increase in nuclei, GFP-transformed A. niger also give an indication of metabolic activity by giving an indication on whether transcription and translation occur.

Splitting up the domains
22.07.2013
Luckily, we got some help from Kal, a really nice guy who offered a lot of help. Several very useful websites he suggested could analyse the AA sequence and give some advice on where is the possible site to split the whole protein. We combined the analysis result with references, we finally got a clear idea (or at least we believe) about the boundary of the domains.
Week 7
July (15.07 - 21.07)

Working on the Wiki
20.07.2013
The design of the wiki is making good progress. Basic layout has been determined and we created icon to indicate different parts of the project.

Article searching
18.07.2013
Totally lost in how to divide each single domain in the lovB gene. By searching the articles, we could find information and experimental details about ACP, KS, MAT and CON domain. While for the others, still virgin land. KR seems to have two subdomains, what the hell~

Database design
16.07.2013
text here
Week 6
July (08.07 - 14.07)
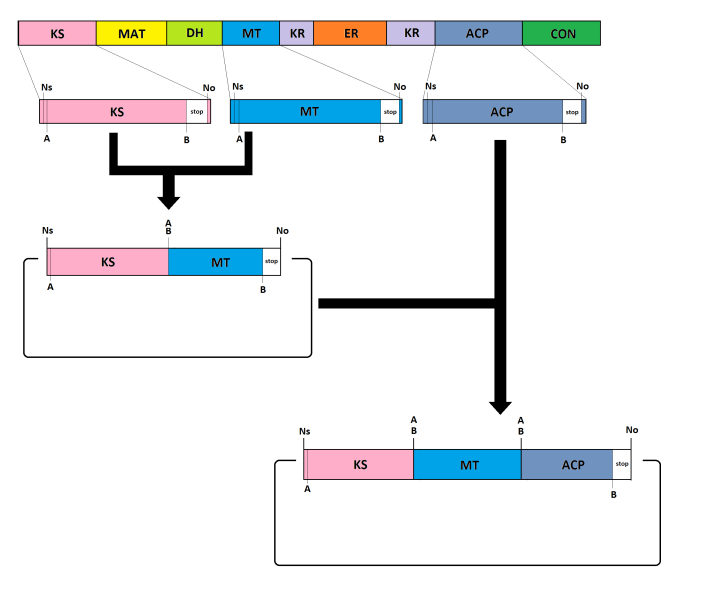
Lovastatin strategy
12.07.2013
Now that we have the general idea the details need to be worked out. Therefore we came up with a strategy to assemble the modules using compatible restriction enzymes. The strategy will enable us to assemble several modules and to have a stop codon at the end of the assembled gene without a frame shift. Also the general planning of the project was worked out this week.

DAPI staining
11.07.2013
It has been found that dormant conidia are predominantly bi-nucleate (85%), the remainder being uni-nucleate. Therefore, if one stains the giant cells with DAPI, which colours the nuclei, one can assess whether the nuclei within the cells are actively dividing. This appears to be the the case and thus indicates that the cells are not in a vegetative state.
Week 1
July (01.07 - 07.07)

Science Café
02.07.2013
We met with the Science Café organisation to discuss the possibilities towards our team organising an evening there with a Biohacker theme.

BioKe meeting in Wageningen
05.07.2013
A BioKe spokesperson came over to Wageningen. We gave her a tour and presentation. We obtained the Q5 PCR kit!

Lovastatin
05.07.2013
Finally the search has begun, this week we have started with literature research for the Lovastatin project. Some topics were for example what is Lovastatin, which genes are involved in Lovastatin production in Aspergillus terreus, and how to measure Lovastatin production. The first idea was to introduce the Lovastatin pathway of A. terreus into A.niger. During the research we found that one of the bigger genes involved is composed of several domains. Our new idea now is to synthesize these domains, making several modules that we can assemble as we wish.
Database planning
01.07.2013
text here
- J.F. Sanchez et al., 2012. Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat. Prod. Rep. Vol. 29, p. 351-371
Week 4
June (24.06 - 30.06)

Calcofluor staining
24.08.2013
text here
Week 3
June (17.06 - 23.06)

Do It Yourself Biohacker Meeting 2
20.06.2013
We had a DIY Biohacker Meeting in the Waag, Amsterdam. The Waag had its grand opening and there was a exposition and party afterwards

Do It Yourself Biohacker Meeting 1
18.06.2013
We had a DIY Biohacker Meeting in the Waag, Amsterdam. Learned how to make your own potato-agar

Finding the right conditions for distinct phenotypes
22.06.2013
As described in literature, N593 formed giant cells after 24h at 44C. Since transcriptome data on N400 from multiple stages in its life cycle is available, this strain is chosen such that this research complements the current RNA landscape profile of A. niger and allows for comparison of data from different life stages. However, unlike N400, N593 repeatedly formed mycelium at 44C. To overcome this effect the temperature was increased to 45C, at which a single cell phenotype was obtained for N593.
Week 2
June (10.06 - 16.06)

Funny group picture
15.06.2013
Opportunity was presented to take a few fun group pictures... we obviously took it

Dinner and discussion
14.06.2013
We met at Mark’s place for dinner and evaluation of possibilities in using secondary metabolites for our project

Last and 8th recruit
13.06.2013
Yeng Ding AKA Danny joind our team. We are now 8 members strong!

ThinkerCell & CloneManager meeting
12.06.2013
The ones from our team who had learned to work with these software tools, during one of the iGem Netherlands courses, taught the others how use it.

Lab supplies
11.06.2013
We ordered reagents for cloning and PCR.

Collaboration
11.06.2013
We contacted the Cornell University iGem 2013 team to talk about possibilities for collaboration. They also work with fungus.

Host Engineering: Research plan
13.06.2013
Different environmental conditions induce changes in phenotypic cellularity of Aspergillus niger. Mapping reads onto the reference genome allows for discovery of patterns in gene expression that are unique to the single cell phenotype.
Week 1
June (03.06 - 09.06)

Host Engineering: Research Question
04.06.2013
Research question:
Finding sets of candidate genes causative to the single cell phenotype in Aspergillus niger by transcriptome analysis.
Subquestions:
- is A. niger metabolically active at 44C?
- does A. niger divide at 44C?
May

Meeting room
26.05.2013
We reserved a room for all future Monday and Thursday lunch meetings in the Forum

Meeting iGem Uppsala
18.05.2013
We met with the iGem Uppsala 2013 team in Uppsala

Created a Gmail account
23.05.2013
We created a Gmail account. Let the spam begin

Created a Twitter account
15.05.2013
We created a Twitter account. Hello world! Meet iGem Wageningen 2013

Chromoprotein sequencing
13.05.2013
Looked into sequencing options: codon optimization to Aspergillus Niger, suffix/prefix, (illegal) restrictionsites. Let’s order some g-blocks!

Lab space
08.05.2013
We arranged our very own lab space!

Host Engineering: why?
20.08.2013
The single-cell phenotype Aspergillus has a higher surface to volume ratio and results in a lower viscosity of the liquid broth, offering perspective on substantially increasing process yields when used in liquid fermentations. Besides from an industrial point of view it is also interesting from an evolutionary perspective, which couples application-oriented research using a directed evolution approach to a more fundamental research topic: the evolution of multicellularity.
April

Good Lab Practice
24.4.2013
We had a lecture on good lab practice and safety in the lab
Task division
18.04.2013
Co-ordinator – Michiel
Secretary – Emiel
Sponsoring - Emiel
Safety - Jingjing
Lab manager – Shreyans
Strategy - Marit
Wiki – Michiel
Human Practice - Marjan

Influence of temperature on germination
20.08.2013
A paper from 1970 shows that at 44C Aspergillus niger does not germinate, but rather forms giant cells. This dimorphism is not unfamiliar within the fungal world, where at lower temperatures the mycelium is formed and at higher temperatures the yeast-like form is found.
Anderson, J. G. and J. E. Smith (1972). "Effects of Elevated-Temperatures on Spore Swelling and Germination in Aspergillus Niger." Canadian Journal of Microbiology 18(3): 289-297.
March
February
January

Title
00.00.2013
Lorem ipsum dolor sit amet, consectetur adipisicing elit, sed do eiusmod tempor incididunt ut labore et dolore magna aliqua. Ut enim ad minim veniam, quis nostrud exercitation ullamco laboris nisi ut aliquip ex ea commodo consequat. Duis aute irure dolor in reprehenderit in voluptate velit esse cillum dolore eu fugiat nulla pariatur. Excepteur sint occaecat cupidatat non proident, sunt in culpa qui officia deserunt mollit anim id est laborum.
javascript
 "
"
