Team:NCTU Formosa/results
From 2013.igem.org
| Line 35: | Line 35: | ||
===sRNA regulation efficiency === | ===sRNA regulation efficiency === | ||
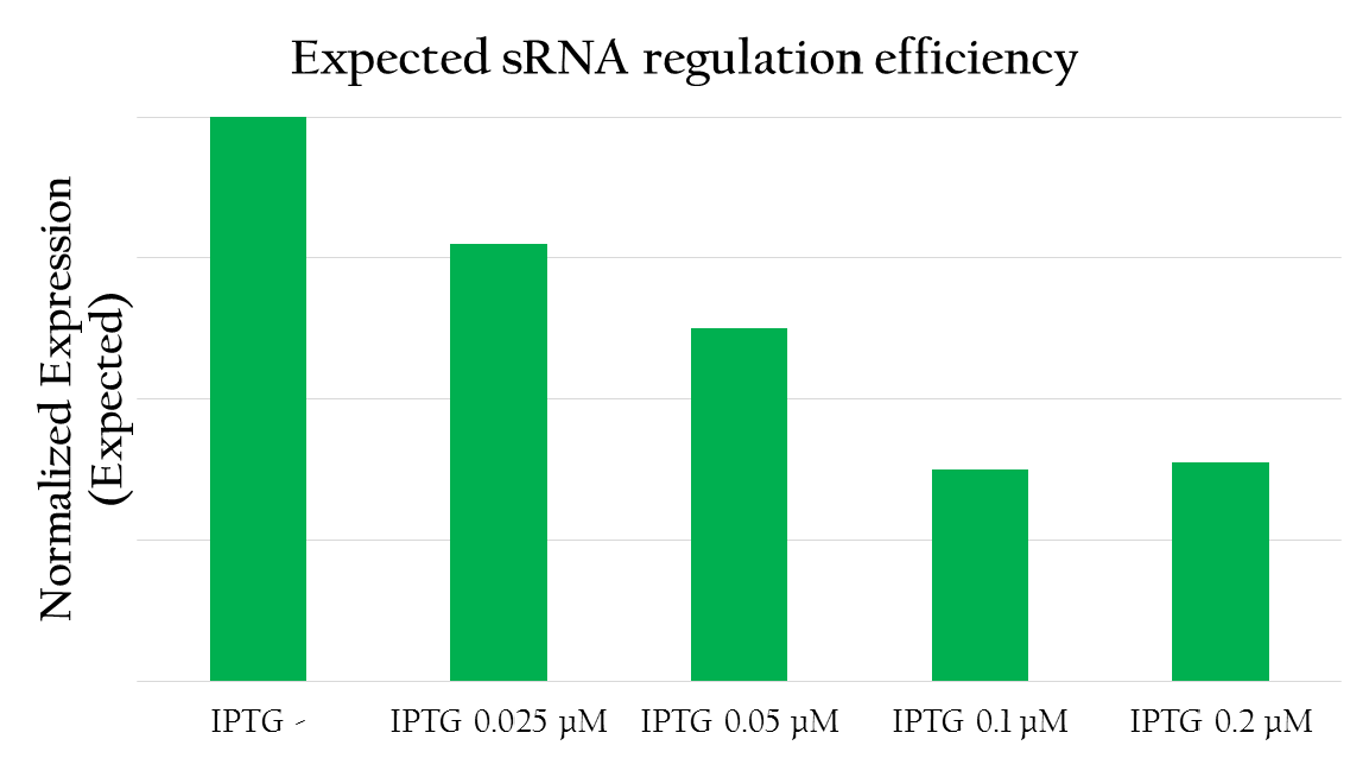
<p>To test the regulation efficiency of sRNA, we employed the following biobricks: Pcons + rRBS + mGFP + J61048 and Pcons + B0030 + lacI + J61048 + Plac + sRNA.</p> | <p>To test the regulation efficiency of sRNA, we employed the following biobricks: Pcons + rRBS + mGFP + J61048 and Pcons + B0030 + lacI + J61048 + Plac + sRNA.</p> | ||
| - | + | [[File:NCTU_sRNA.JPG|center|600px|Fig 4. sRNA regulation]] | |
<span class="undetermined">The graph above shows the relationship between the concentration of IPTG added and the red florescent measured. The more IPTG added, the more sRNA can be produced by Plac as IPTG serves to activate the promoter. In other words, the amount of sRNA and the concentration of IPTG forms a liner relationship. With more IPTG, and therefore, more sRNA the amount of red fluorescent measured decreases. This implies that sRNA can effectively regulate the expression of RFP by decreasing the translation efficiency and the stability of rRBS. </span> | <span class="undetermined">The graph above shows the relationship between the concentration of IPTG added and the red florescent measured. The more IPTG added, the more sRNA can be produced by Plac as IPTG serves to activate the promoter. In other words, the amount of sRNA and the concentration of IPTG forms a liner relationship. With more IPTG, and therefore, more sRNA the amount of red fluorescent measured decreases. This implies that sRNA can effectively regulate the expression of RFP by decreasing the translation efficiency and the stability of rRBS. </span> | ||
Revision as of 09:46, 21 September 2013
The current progress of our project, including detailed information of the experimental data and the overall evaluation of the practicability of this project.
Contents |
RBS efficiency
We used the following biobrick to test the efficiency of different ribosome binding sites:
- Pcons+BBa_B0034+mRFP+Ter
- Pcons+[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1017202 BBa_K1017202]+mRFP+Ter
- Pcons+BBa_B0030+mRFP+Ter
- Pcons+BBa_B0032+mRFP+Ter
- control:pet40
The control is pet40 and the reporter gene is mRFP, Figure 1 and Figure 2 show the difference of color.
(undetermined)
37 degrees Celsius RBS
Using biobrick, Pcons+37°C RBS+mGFP+J61048, we tested the function of the 37°C RBS under different temperatures.
As shown on Fig 3. the expression of mGFP under 37°C and 42°C reached a higher OD value in less time than the expression below 37°C. This demonstrates the fact that 37°C RBS functions more efficiently above or at 37°C. On the other hand, the fact that 37°C RBS leads to moderate expression even under 37°C degree might be a problem, since then we cannot make sure that only single gene is expressed under 37°C.
sRNA regulation efficiency
To test the regulation efficiency of sRNA, we employed the following biobricks: Pcons + rRBS + mGFP + J61048 and Pcons + B0030 + lacI + J61048 + Plac + sRNA.
The graph above shows the relationship between the concentration of IPTG added and the red florescent measured. The more IPTG added, the more sRNA can be produced by Plac as IPTG serves to activate the promoter. In other words, the amount of sRNA and the concentration of IPTG forms a liner relationship. With more IPTG, and therefore, more sRNA the amount of red fluorescent measured decreases. This implies that sRNA can effectively regulate the expression of RFP by decreasing the translation efficiency and the stability of rRBS.
 "
"