Team:BostonU/Data
From 2013.igem.org
| Line 57: | Line 57: | ||
</html> | </html> | ||
| - | + | ||
[[File:BU_flow_main.png]] | [[File:BU_flow_main.png]] | ||
Revision as of 03:52, 26 September 2013
Data Collected
New Part Creation
Characterization of Level 1 MoClo Devices
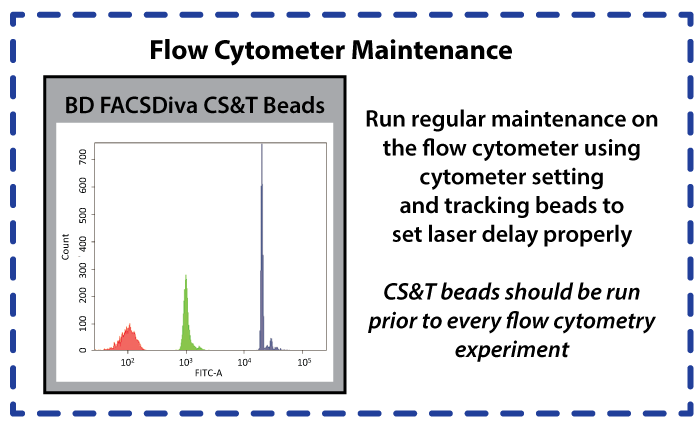
The Cytometer Setup and Tracking beads offered by BD biosciences are utilized to set the laser delay and optimize the cytometer settings prior to running any samples through the Fortessa.
We also utilize an external positive control in the form of Spherotech's 8-peak particles (cat number RCP-30-5A), which fluorescence when run through all of the Fortessa laser/filter combinations we are using this summer.

Datasheet Tool
The Datasheet Tool was coded primarily in Java for the back-end server side and in HTML from the front-end client side. The app was coded in the NetBeans IDE and we stored all of our code on a private GitHub repository.
Eugene Results
We wrote Eugene scripts for our projects this summer. The wiki upload option does not allow .eug Eugene files so we uploaded our code as plain text files. To run our code in the Eugene Scriptor, simply save the code as .eug.
The pConstitutive Library Eugene file shows all of the constitutive promoter and fluorescent protein permutations using the Anderson promoter library.
The pRepressible Library Eugene file shows all of the possible repressible promoter and gene relationships based on our MoClo library.
The Quorum Sensing Eugene file shows the permutations possible with the specified rules, such as the repressing relationship between certain promoters and genes and the small molecule arabinose inducing the pBad promoter.
 "
"