Team:DTU-Denmark/Protein Models
From 2013.igem.org
(Difference between revisions)
| Line 4: | Line 4: | ||
==Mutant 1== | ==Mutant 1== | ||
| - | ==AMO== | + | ===AMO=== |
Ammonia monooxygenase | Ammonia monooxygenase | ||
[[File:DTU_AMO.png|640px|center]] | [[File:DTU_AMO.png|640px|center]] | ||
| + | |||
| + | ''homology models of all three subunits of AMO'' | ||
===Hao=== | ===Hao=== | ||
Hydroxylamine oxidoreductase | Hydroxylamine oxidoreductase | ||
Revision as of 19:23, 1 October 2013
Protein Models
It turns out that the structures of some of the proteins we want our mutants to express have been determined.
Contents |
Mutant 1
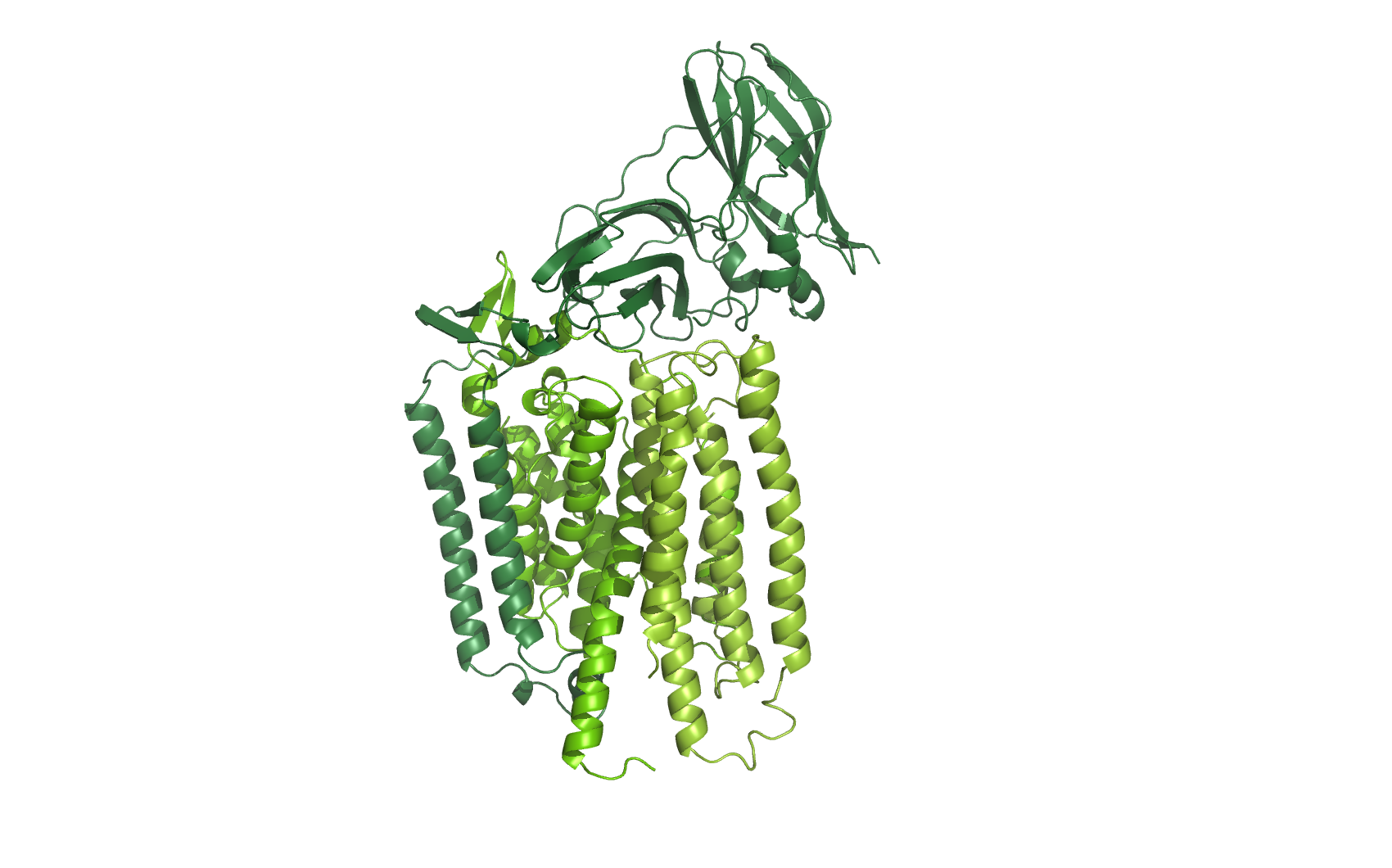
AMO
Ammonia monooxygenase
homology models of all three subunits of AMO
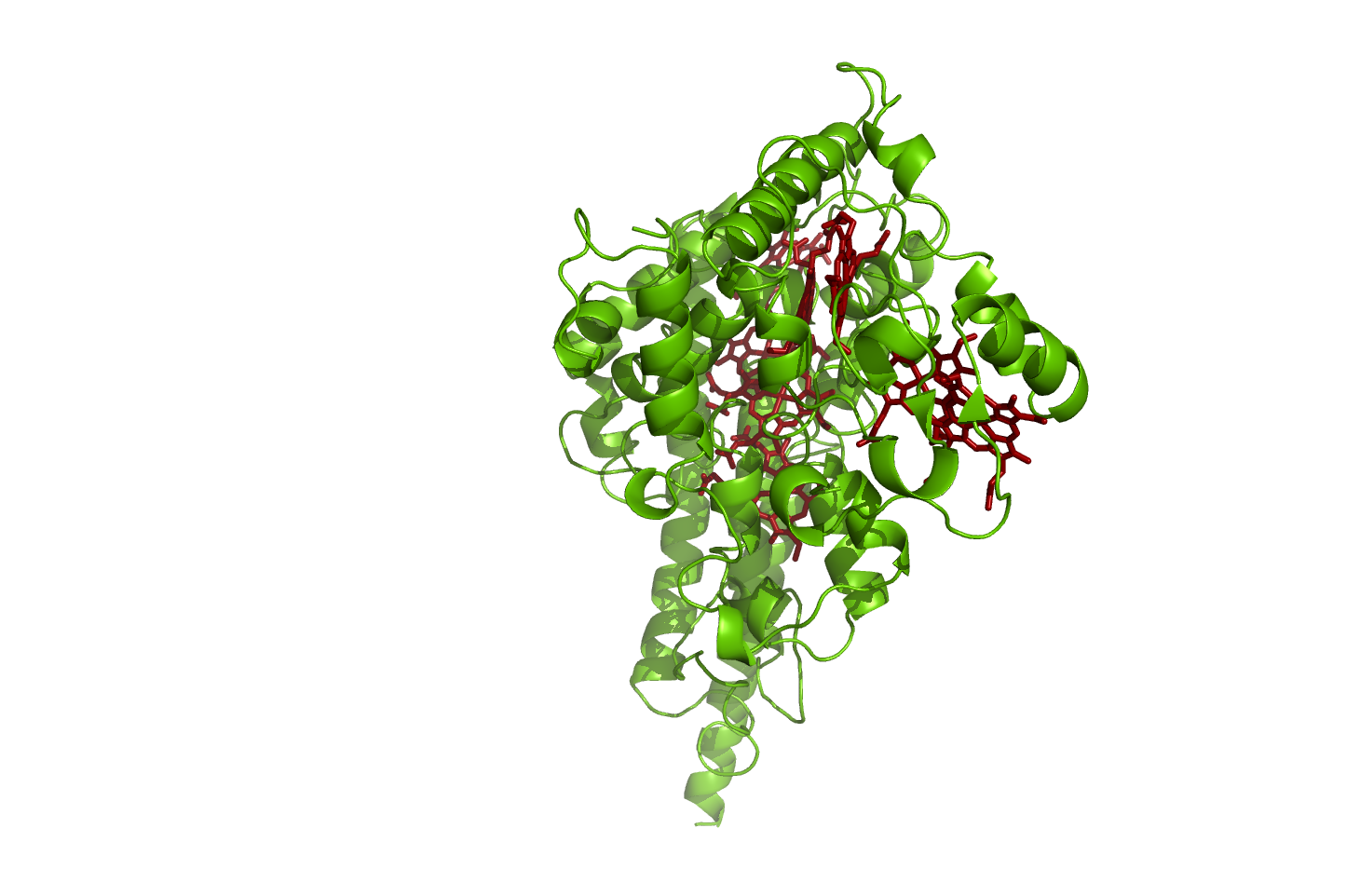
Hao
Hydroxylamine oxidoreductase
source [http://www.rcsb.org/pdb/explore.do?structureId=1fgj 1FGJ]
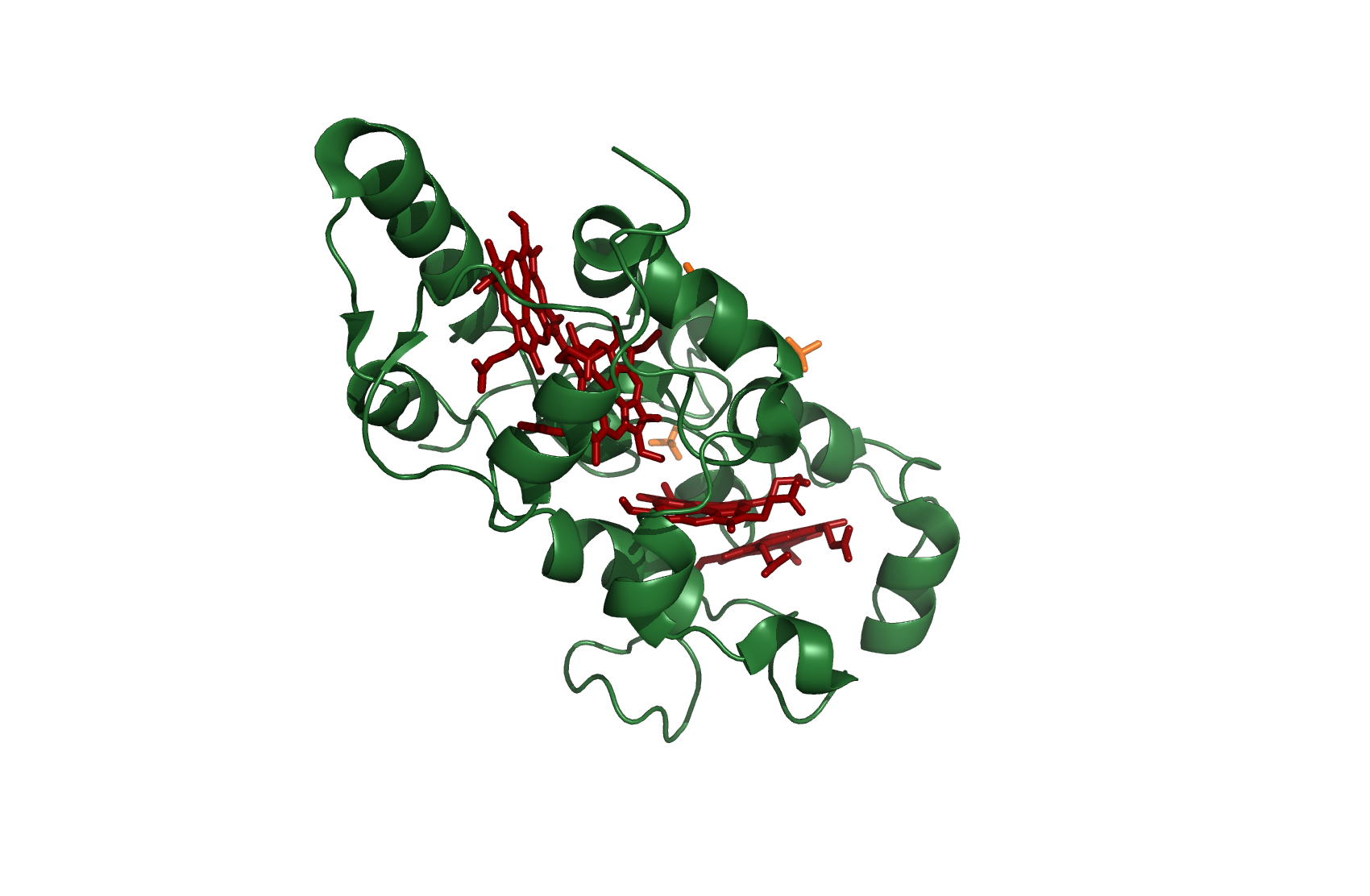
cc554
Cytochrome c554
source [http://www.rcsb.org/pdb/explore.do?structureId=1ft5 1FT5]
Mutant 2
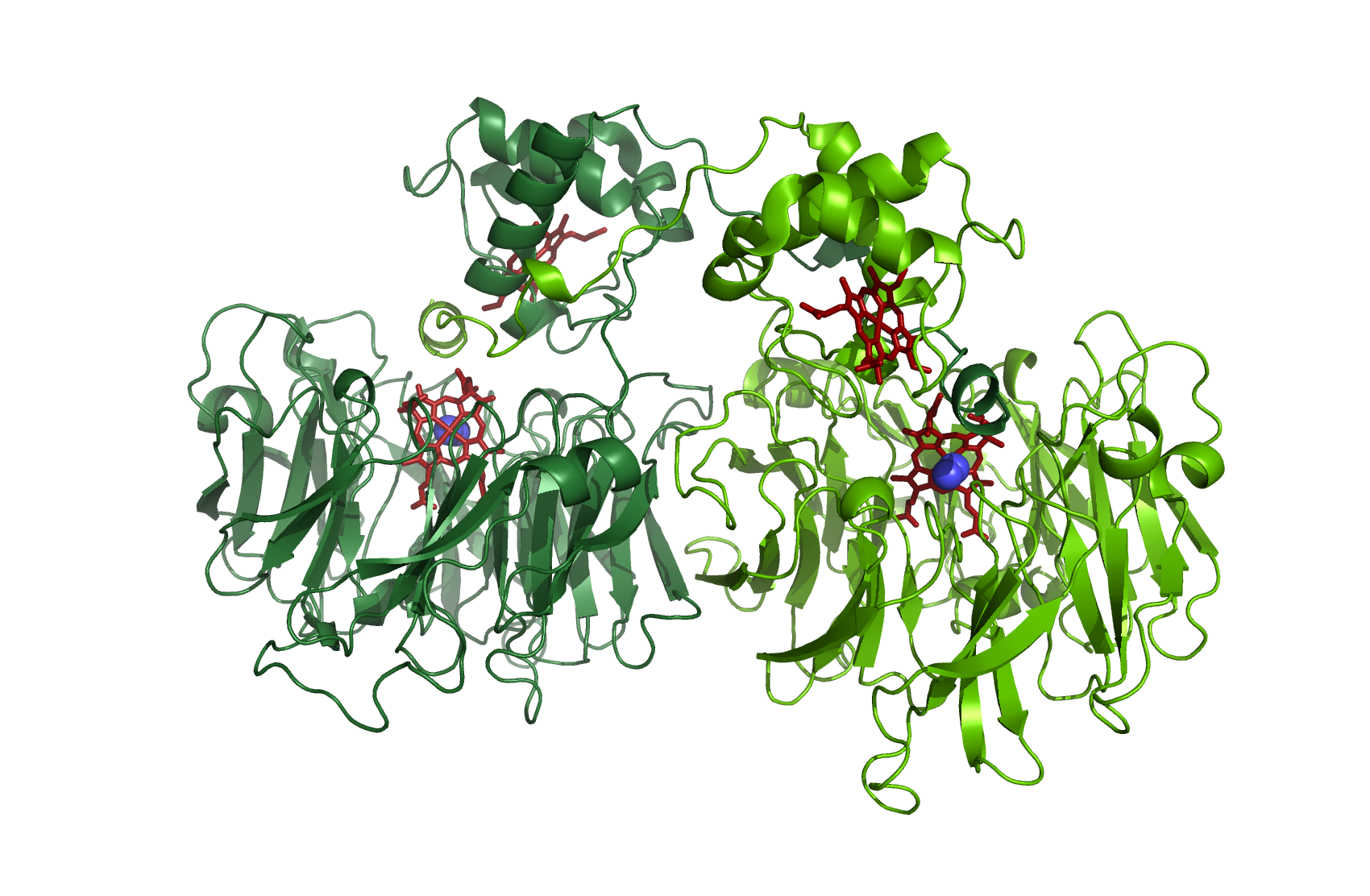
NirS
Nitrite reductase
As a dimer with N2 - source [http://www.rcsb.org/pdb/explore/explore.do?structureId=1nno 1NNO]
All models were made and visualized in [http://sourceforge.net/projects/pymol/ PyMOL v1.6]
 "
"