Team:HokkaidoU Japan/Overview/Background
From 2013.igem.org
| Line 88: | Line 88: | ||
<dt>RBS Selector</dt><dd>RBS Selector is for adjustment of multiple expressions, like complex metabolisms.</dd> | <dt>RBS Selector</dt><dd>RBS Selector is for adjustment of multiple expressions, like complex metabolisms.</dd> | ||
</dl> | </dl> | ||
| - | We named these two devices ''Maestro <span class="italic">E.coli</span>'' Random Operon Shuffling Kit! As a maestro harmonizes music, let's harmonize proteins in the bacteria! | + | We named these two devices ''Maestro <span class="italic">E. coli</span>'' Random Operon Shuffling Kit! As a maestro harmonizes music, let's harmonize proteins in the bacteria! |
</p> | </p> | ||
Revision as of 02:27, 29 October 2013
Maestro E.coli
Overview

Background
Synthetic biology is one of the most interesting fields in 21st century. Its goal is to comprehend and reproduce the marvels of living things. As members of syn-bio community, a lot of iGEMers have found interesting proteins. However, the focuses of these projects are “qualities” of proteins. As well as the uniqueness of proteins, the expression levels of proteins contribute to the marvelous functions of living things. The next goal of syn-bio is to control “quantities” of proteins.
To control protein quantities in the organism, we must control transcription, a step from DNA to mRNA and translation, a step from mRNA to protein. Transcription is regulated by promoter region. Translation is regulated by ribosome binding site: RBS. Depending on promoters, the transcription rate varies about 1000 fold. Similarly, depending on RBSs, the translation rate about 100 fold. So, to control quantity, it is essential to figure out the characteristics of these expression-regulatory regions and adjust them.
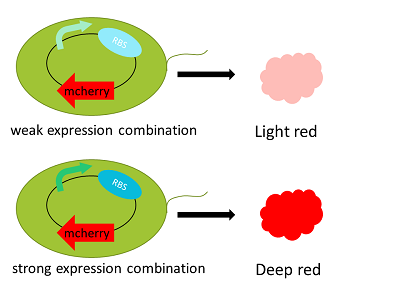

Recently, many studies have been done to theoretically predict the amount of gene expression by the sequences of expression-regulatory regions. There are some accurate in vivo expression efficiency predictions. However, even if you were able to predict it exactly, you could not adjust it to an optimal level because you don't know the best rate for the bacteria.
Suppose you are planning to express beneficial product, like antibodies. You may think that the strongest promoter and RBS lead the maximal yield of the proteins. But actually, selecting the strongest one isn't always the best strategy. Overexpressed proteins can be perceived as detrimental by bacterial immune system and packed into inclusion bodies.

Another example is a multiple expression, such as mixing color of chromoprotein. Theoretically, three different chromoproteins can express any kind of colors by fixing each expression level. But this adjustment is too sensitive to be made instinctively.
Furthermore, the fact that sequence of CDS may affect transcription or translation efficiency is known from some studies. It means the strength of promoter or RBS could be changed by CDS.

Over these challenging problems in syn-bio, we put forward one solution like a Columbus's egg. "No more prediction, let's experiment!" ---- that is, we should try several promoters or RBSs and select the best one or the best combination. We don't know the best answer, but bacteria do.
This is surely the best way, but trying every pattern costs a lot of time and labor. We overcame this obstacle by using a one-pot DNA shuffling method, namely, "Golden Gate Assembly". And then, we made two kits to optimize transcription and translation. For these kits, we made artificial promoter and RBS families. They are created based on concrete philosophies and characterized well. Please refer Promoter and RBS for details.

We named these kits "Promoter Selector" and "RBS Selector".
- Promoter Selector
- Promoter Selector is for adjustment of relatively simple expression, like a production of antibodies reffered above.
- RBS Selector
- RBS Selector is for adjustment of multiple expressions, like complex metabolisms.
 "
"


