Team:Tianjin/Project/Introduction
Contents |
Background
Unsustainable energy demands worldwide and the acceleration of global climate change have produced a formidable scientific research problem. To figure out this problem, scientist have put great attention on biofuels, hoping to find a solution to energy crisis and climate change. Among all biofuel, alkanes which are the predominant components of gasoline, diesel, and jet fuels, may be an ideal candidate for biofuel.[1] Nowadays, many pathways of alkanes biosynthesis has been reported, and it is possible for microorganisms to transform renewable resources such as lignocellulosic biomass into advanced biofuel under the help of the technology of synthetic biology.
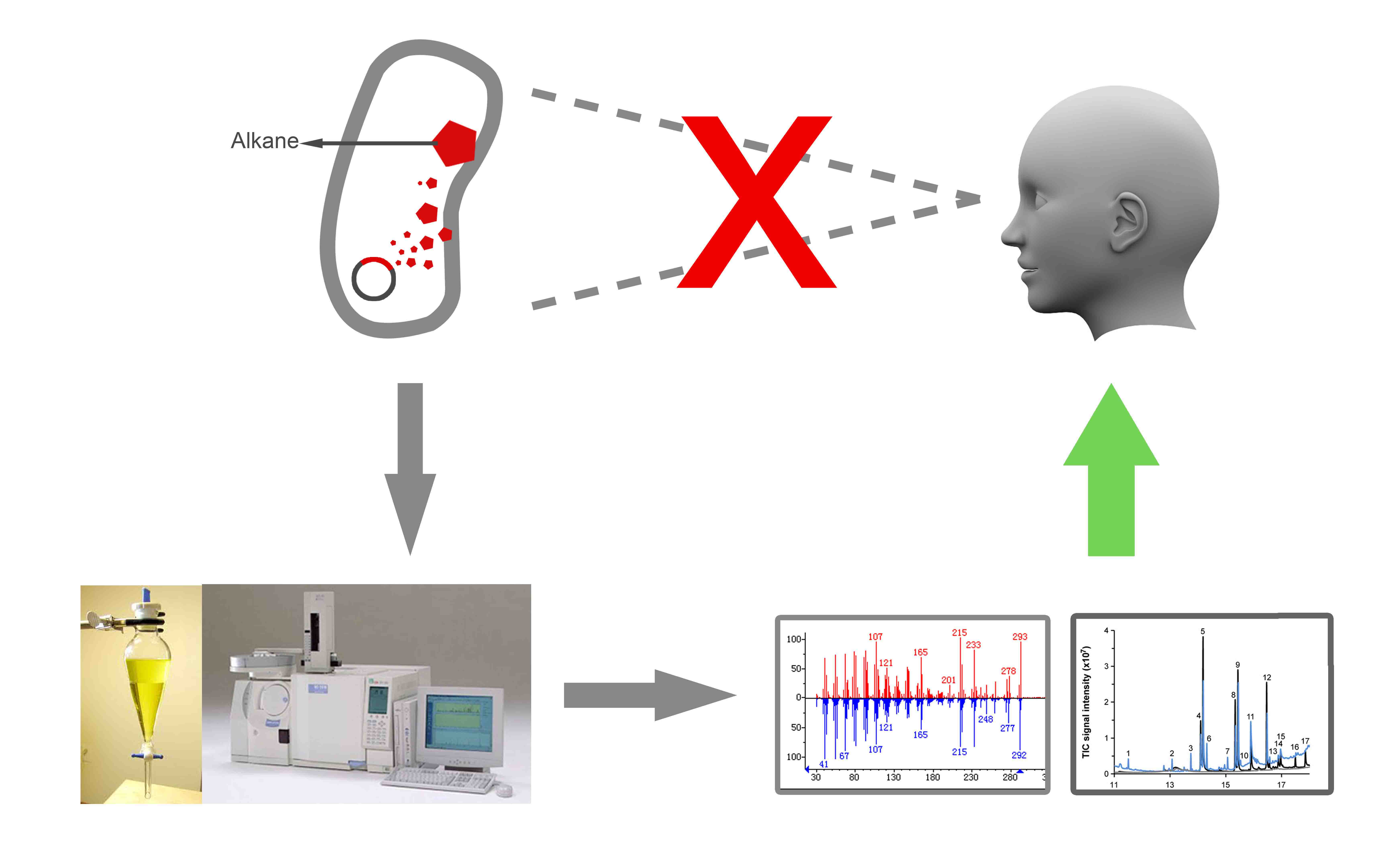
However, it is a long way to engineer a microorganism to produce alkanes and to optimize its productivity to satisfy the needs of industrial process. For researchers, it is important to detect and measure the concentration of alkanes, especially real time concentration of alkanes in vivo. These information are very helpful for researchers to engineer and increase the productivity of biofuels. Unfortunately, unlike carotene, violacein and lycopene which are naturally coloured and easily detected, alkanes are inconspicuous small molecules, without any property for human beings to detect its existence. The measurement of alkanes mainly depends on high-tech equipments such as gas chromatography and mass spectrometry. On the other hand, all the samples needed special treatment before detecting alkane, for the in vivo alkane, it is needed to extract alkanes from the cell by using ethyl acetate. So the current method can hardly perform a real-time in vivoItalic text detection of alkanes.
In a word, detection and measurement of alkane is a complex process and needs high-tech equipment. An easy method of alkane detection may be of great help.
Inspiration
To solve the problem, our project intends to construct an alkane bio-sensing device, to transform the signal of alkane concentration into signals which can be easily detected such as chromophoric or florescent signals. We hope this device can help us “mark” the existence of alkane and detect them. For the engineered strains with this device, people can detect its production of alkane in vivo easily and quickly.
The key of constructing this device is to find a genetic system related to alkane sensing. The oil-degrating microorganisms such as P. putida Gpo1, Alcanivorax borkumensis SK2 and Acinetobacter baylyi ADP1, caught our attention. These microorganisms can use alkanes as the sole carbon source to support growth, and the expression of genes related to degrading alkane is regulated by special system, to make sure the expression of these genes is only activated in the presence of alkanes. Among these regulation systems, we choose AlkR-PalkM system from Acinetobacter baylyi ADP1 as the core of our alkane biosensing device. In this system, the transcriptional regulatory protein AlkR can form a dimer and recognize the existence of alkane. This complex can activate the promoter of alkM which encodes alkane hydroxylase, and promote the metabolization of alkanes. Compared with other alkane degrading regulation system such as AlkS-PalkB from P. putida Gpo1 and A. borkumensis SK2, AlkR-PalkM has a very special advantage: this system cannot be activated by alkanol which is the main byproduct of alkane biosynthesis. (Alkanol can activate AlkS-PalkB system) So AlkR-PalkM system can reflect the alkane productivity of engineered strains more accurately.
We hope that the gene circuit AlkR-PalkM-bioreporter will help us detect and measure the alkanes produced by our engineered E. coli more easily and quickly.
Reference
[1] Yan Kung, Weerawat Runguphan, and Jay D. Keasling. “From Fields to Fuels: Recent Advances in the Microbial Production of Biofuels.” ACS Synth. Biol. 2012, 1, 498−513
[2] Mathew A Rude and Andreas Schirmer.“New microbial fuels: a biotech perspective.” Current Opinion in Microbiology 2009, 12:274–281
[3] Andreas Schirmer et al. “Microbial Biosynthesis of Alkanes.” Science 2010: Vol. 329 no. 5991 pp. 559-562
[4] Rebecca M Lennen and Brian F Pfleger “Microbial production of fatty acid-derived fuels and chemicals” Current Opinion in Biotechnology 2013, 21:1–10
[5] M. Kalim Akhtar, Nicholas J. Turner, and Patrik R. Jones. “Carboxylic acid reductase is a versatile enzyme for the conversion of fatty acids into fuels and chemical commodities.” PNAS January 2, 2013,vol. 110, no. 1, 87–92
[6] Rekha Kumari, Robin Tecon, Siham Beggah et al.(2011) “Development of bioreporter assays for the detection of bioavailability of long-chain alkanes based on the marine bacterium Alcanivorax borkumensis strain SK2.” Environmental Microbiology 13(10), 2808–2819
[7] Zhang, Dayi, et al.(2012) "Whole-cell bacterial bioreporter for actively searching and sensing of alkanes and oil spills." Microbial Biotechnology 5.1: 87-97
Supplement Information
a.More information about biofuel and alkane
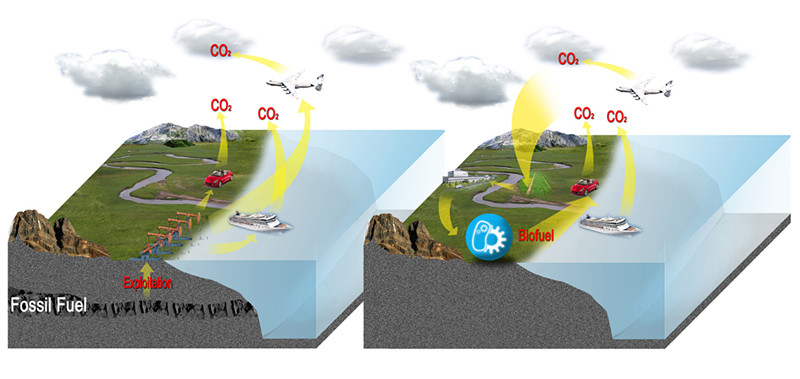
Biofuels are non-fossil fuels produced from solar energy that is chemically stored as high-energy organic compounds by organisms. Atmospheric CO2 is first fixed into sugars by photosynthesis. The sugars can then be transformed into fuel compounds by engineered microorganisms, and burning of these biofuels drives an engine, re-releasing CO2 back into the atmosphere. This closed CO2 cycle is driven by the energy of the sun and constitutes a more carbon-neutral process than the direct burning of fossil fuels, the cause of ever-increasing CO2 levels.
Among all kinds of biofuels, alkanes have several advantages compared with other biofuels. First, alkanes have higher energy density, for example, enthalpy of combustion of pentadecane is approximately -47.0 MJ/kg compared with -29.7 MJ/kg for ethanol [2]. Second, compared with ethanol, the first generation biofuel, with a freezing point of -114℃, alkanes have a higher freezing point of about -3~19℃, so they are more likely to be compatible with existing engines as well as transport and storage infrastructures. Besides, they can serve as drop-in replacement for fossil fuels.
b.More information about alkane biosynthesis pathway
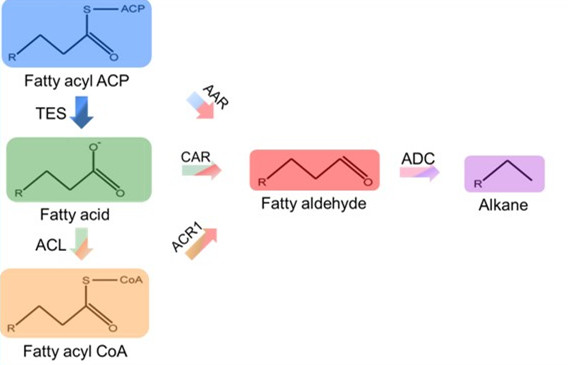
Pathways of long-chain alkane sythesis in microbes has been studied. Two enzymes, acyl-ACP reductase(AAR) and aldehyde decarbonylase(ADC), were heterologously expressed in E.coli to reduce fatty acyl-ACPs to corresponding aldehydes and then convert them to alkanes[3]. Fatty aldehydes can also be produced from fatty acids and fatty acyl-CoAs, catalyzed by carboxylic acid reductase(CAR) and acyl-CoA reductases(ACR) respectively, which has been identified in many species[4,5].
c.More information about AlkR-PalkM
Protein ALKR and promoter alkM are originally found from Acinetobacter baylyi ADP1. The ALKR-PalkM gene circus is part of the regulation system of alkane metabolism.
Acinetobacter sp. are ubiquitous bacteria in natural aquatic and soil environment that are frequently found to be capable to degrade a broad range of carbon chain alkenes and alkanes[3]. And Acinetobacter baylyi ADP1 is able to use long-chain-length alkanes with at least 12 carbon atoms as the sole source of carbon and energy[4].
In Acinetobacter baylyi ADP1, P-alkM is the promoter of alkane hydroxylase genes which encodes alkane hydroxylase(alkM), a crucial enzyme in the degradation of alkanes. AlkM, together with rubredoxin (RubA) and rubredoxin reductase(RubB) forms a three-component alkane monooxygenase complex which oxidate inert alkane to the respective primary alcohol[4].
AlkR is an AraC/XylS-like transcriptional regulatory protein. It includes C-terminal DNA-binding domain for promoter binding and N-terminal domain for inducer recognition[2]. Its function is to recognize alkane molecule after its own dimerization and induce promoter alkM. The activity of alkane hydroxylase could impede ALKR’s function. When alkane molecules occur, they could be recognized by protein ALKR. A three component complex is formed. The inducer complex can bind with promoter alkM and activate the genes in the downstream of PalkM.
 "
"