Team:NCTU Formosa/project
From 2013.igem.org
A multiple regulated-system was built using three different regulation mechanisms including red light, temperature, and sRNA. In other words, it is multitasking genetic engineered machine that can express a variable genes depending on the different command given.
Contents |
Introduction
It is not a dream any more that bacteria engineered machines give us the opportunity to fulfill our mission and so, we thought that one day, we could demand bacteria to work specifically along with different transition of environmental conditions. Our basic concept is ,“Each work works under a specific condition”. We use three regulating factors, light, temperature and sRNA to design a model that is cross-regulated and is able to work under different given environmental conditions. Through pairing specific conditions, we will only have to change different reacting conditions for turning on or shutting down reactions, furthermore, we use the sRNA inhibiting system to turn off unnecessary reactions. With noninvasive regulators and an easy way to adjust, such system controlled by multiple regulators is believed to be used in different applications that require specific control.
sRNA
Principle and Why we use it
(undetermined)
Design
Small RNAs (sRNAs) have become increasingly significant in playing the role of bacterial gene regulation. Most sRNAs interact with the targeted mRNAs by imperfect base pairing, reducing the translation efficiency and recruiting chaperones such as Hfq for translation termination. In other words, sRNAs regulate gene expression by forestalling translation. It is an effective and specific regulated-system that functions under RNA level. Since this regulated-system has already been employed in vivo, it is necessarily to design artificial sRNA that targets specifically to the desired genes. This way, we can prevent the sRNA from effecting undesired genes.
Unlike the regulated-system that popular used in iGEM project(e.g., tet promoter & tetR, Lux promoter & LuxR, etc.), sRNA regulated-system seems more efficient. Because the inhibition works under RNA level, which means the energy waste in E.coli is less than other system(producing proteins).
The sRNA employed in this project was picked from a library of artificial sRNA that was constructed by fusing a randomized antisense domain of Spot42, the scaffold that is known to recruit the RNA chaperons. The sRNA we picked contains a consensus sequence, 5’-CCCUC-3’, that can base pair with the SD sequence due to complementarity. This sRNA, as expected, effectively regulates gene expression by reducing translation efficiency and recruiting Hfq. In addition, this sRNA shows high specificity against its targeted gene, ompF, as it doesn’t hold significant activity against other genes from E. Coli genome (Sharma and others, 2011).
The sRNA picked is competent, but we hoped that it can target any desired gene. With that said, we designed a RBS by employing the sRNA targeting region from ompF and making the AUG codon sufficiently apart from the SD sequence for ribosome binding. By adding this RBS to the upstream of any desired gene, the gene can be regulated by sRNA.
37 degree Celsius RBS
Principle and Why we use it
(unfinished)
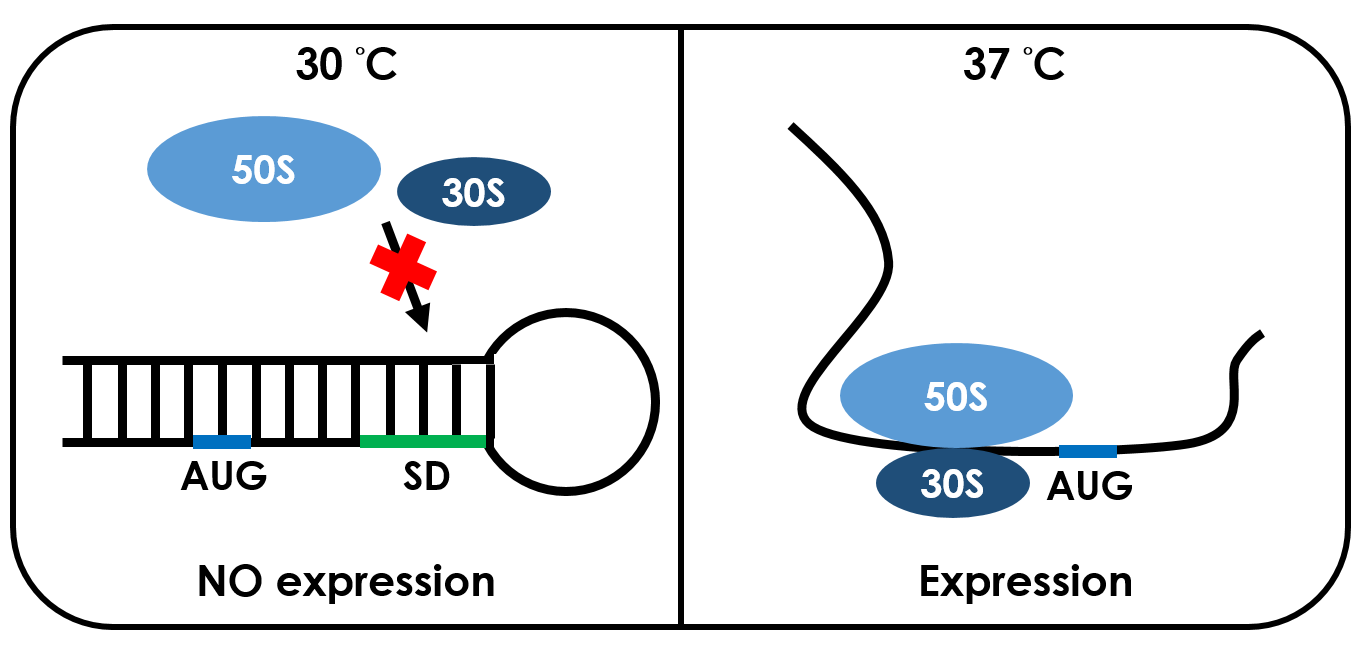
An RNA thermometer is a temperature-sensitive non-coding RNA molecule which regulates gene expression. The expression of heat-shock, cold-shock and some virulence genes are coordinated in response to temperature changes.RNA thermometers are thermo-sensors that regulate gene expression by temperature-induced changes in RNA conformation.There are several systems suggested in literature that are based on RNA secondary structure.This structural transition can then expose or occlude important regions of RNA such as a ribosome binding site, which then affects the translation rate of a nearby protein-coding gene.So we can say that the RNA thermometers are just like riboswitches. Apart from protein-mediated transcriptional control mechanisms, translational control by RNA thermometers is a widely used regulatory strategy.
Unlike normal RBS, the special RBS has a unique hairpin structure if the temperature drops below a certain temperature, the RNA will form stable base-pairs on the Shine-Dalgarno sequence, disabling the ribosome to bind. The base-pairing of this RNA region will block the expression of the protein encoded inside it .And since the structure is sustained by base pairing, heat can be employed to break the hydrogen bonds. With the bonds broken, the hairpin would be unfolded, causing the SD site to be exposed and available for ribosome binding. This means that by raising temperature to a certain threshold, the special RBS can function as a normal RBS.In this way gene expression can be regulated on the RNA level by temperature.
Design
(undetermined)
Light control
mechanism1
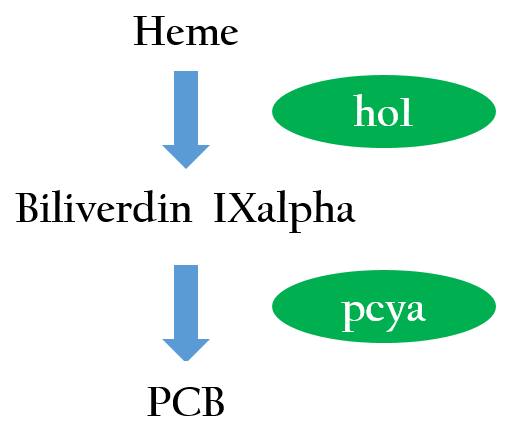
We want to create a light sensor in Escherichia coli, but Photoreceptors are not found in E.coli. Therefore, we introduced two phycocyanobilin-biosynthesis genes (ho12 and pcyA3) from Synechocystis to E.coli. Figure 3 depicts the phycocyanobilin-biosynthesis pathway.
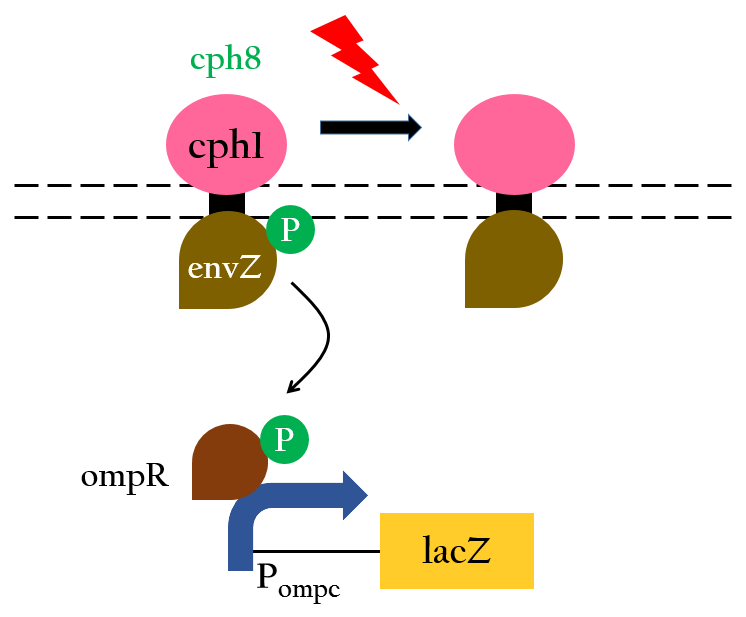
The chimera Cph8 is consist of the phytochrome Cph1 and the histidine kinase domain and response-regulator from EnvZ-OmpR. Cph8 autophosphorlates ompR, then phosphorylated-ompR activates the ompC promoter and turn on the downstream gene.
Red light inhibits the autophosphorylation of Cph8 and turns off the gene expression. Figure 4 depicts this mechnism.
Pred promoter
Why we use it
(undetermined)
Design
(unfinished)
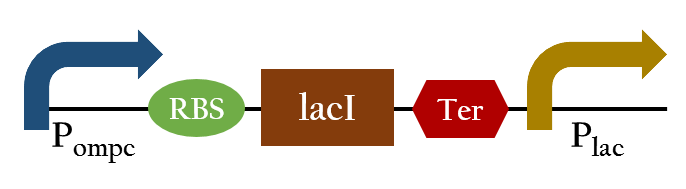
Upon exposure of red light, the phosphorylation activity is hindered, and therefore, the promoter remains repressed and the downstream genes cannot be translated. Such negative control, however, is more complicated and less straightforward than a simple positive control. With that said, we converted negative control of Pompc into positive control by building a biobrick called Pred.
In the presence of red light, Pompc is shut down and lacI cannot be expressed to repress Plac. As a result, Plac is activated and all the downstream genes can be expressed. In the absence of red light, whatsoever, Pompc is activated as phosphorylation is triggered and lacI is expressed. Consequently, lacI respressed Plac and the genes downstream cannot be expressed. By employing Pred, we can achieve positive control using red light,
Reference
- Levskaya, A. et al (2005). Engineering Escherichia coli to see light. Nature, 438(7067), 442.
- part BBa_I15008;MIT Registry of Standard Biological Parts
- part BBa_I15009;MIT Registry of Standard Biological Parts
Mechanism of light induced-device
Pathway Introduction
This is the pathway we designed by using the three different regulating factors. We tend to set up four conditions for the E.coli to work by paring the light and the temperature controls. Distinguished by whether red light exists or not, the pathway divided into two parts, the Red Light Induced Circuit and the Dark Response Circuit. We use four different fluorescent proteins as reporter genes to test that whether the switch of our pathway is successful.
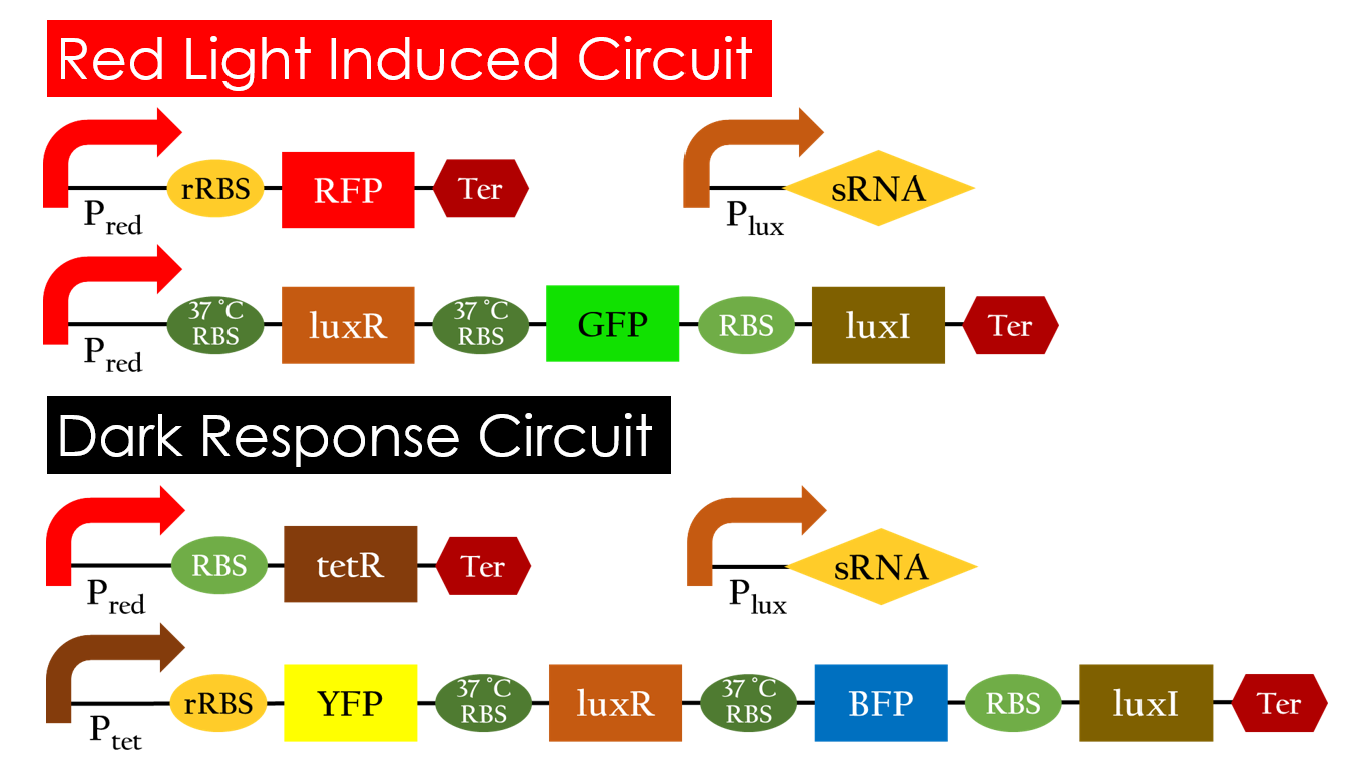
Red Light Induced Circuit
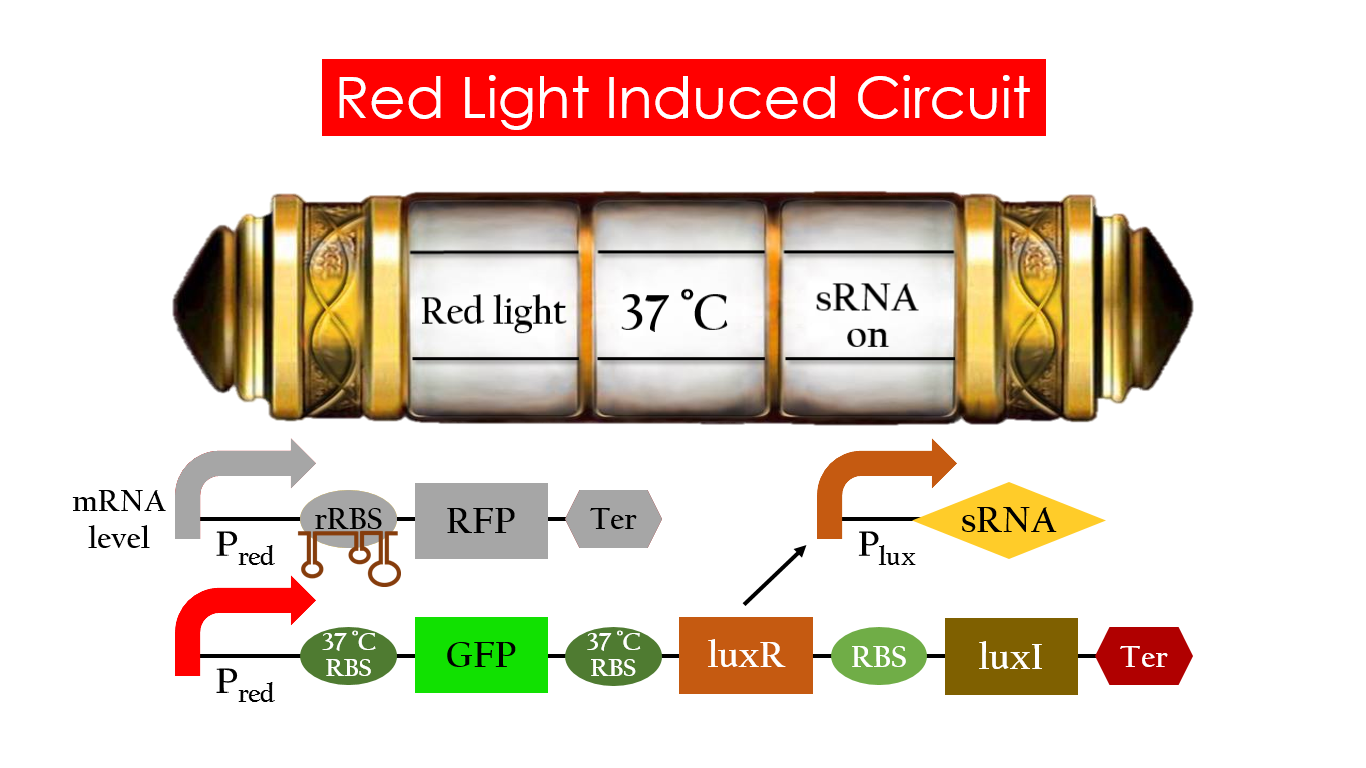
At 30°C and under red light, Pred is activated for translation proceeds. However, ribosomes can only bind to the normal RBS, as 37°C RBS (tRBS) forms a hairpin structure, forestalling ribosomes from binding. The only gene downstream of the normal RBS is RFP, so only RFP would be expressed.
On the other hand, with red light, 37°C RBS unfolds at 37°C , resulting in the expression of both LuxR and GFP. LuxR would binds with AHL to form a complex that activates Plux. The activation of Plux produces sRNA that binds to the normal RBS on mRNA level, blocking it from ribosomes. As a result, RFP would not be expressed and only GFP remains at 37°C .
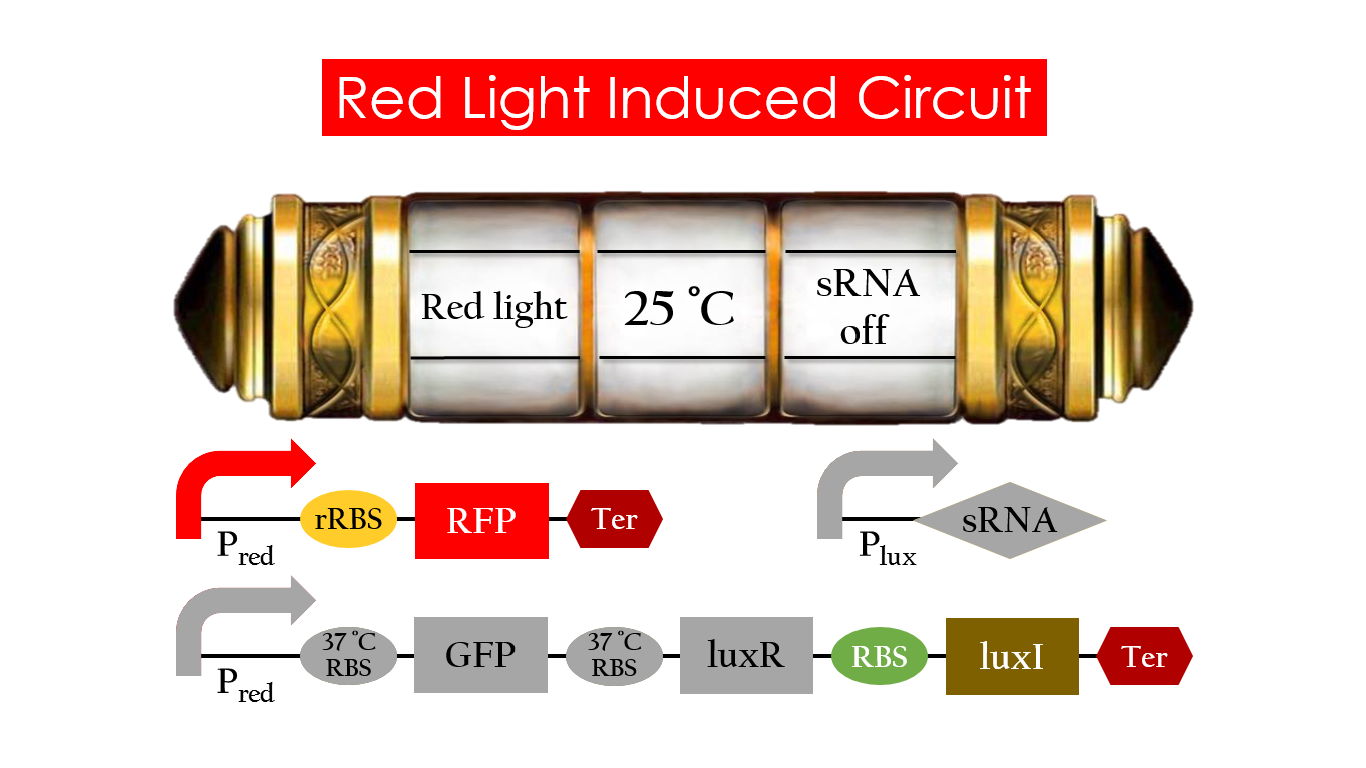
Without red light, Pred would not be activated, and therefore, the red light induced circuit is completely shut down in the dark.
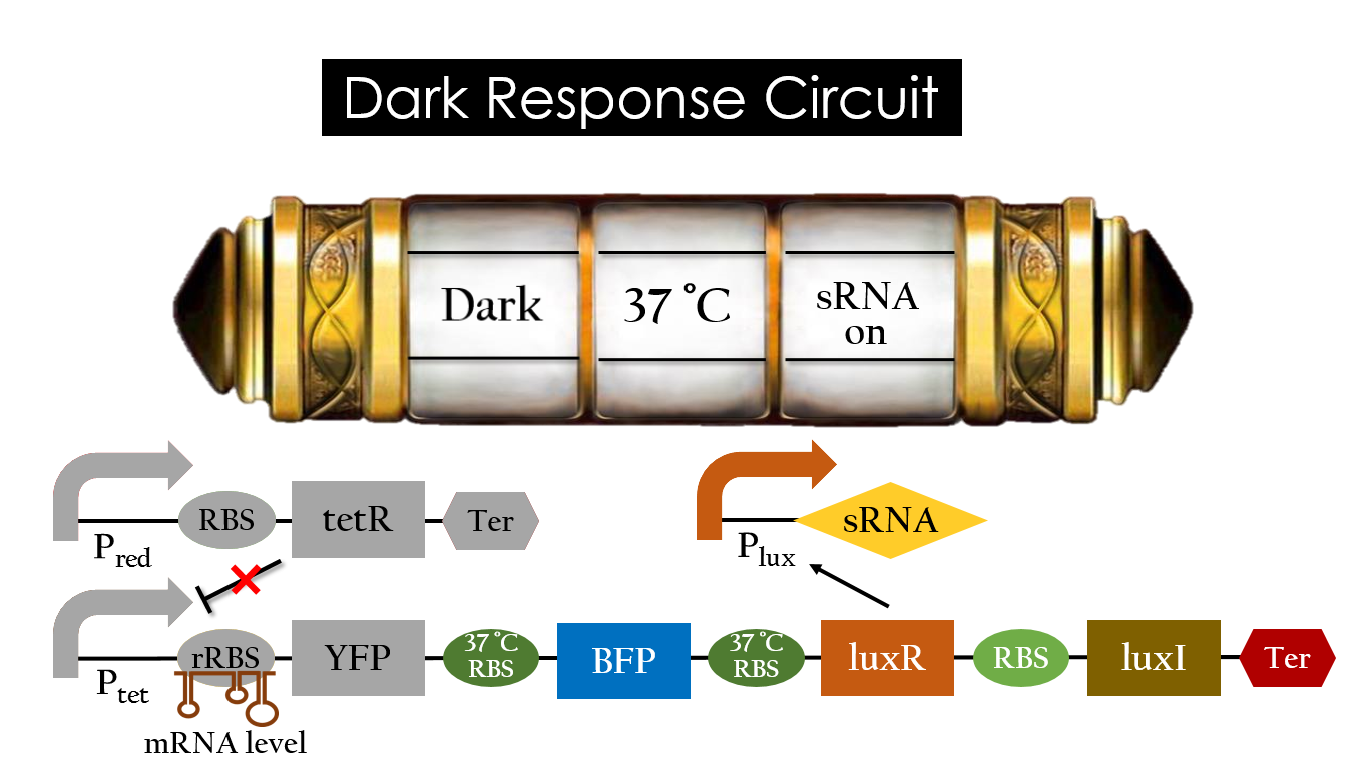
Dark Response Circuit
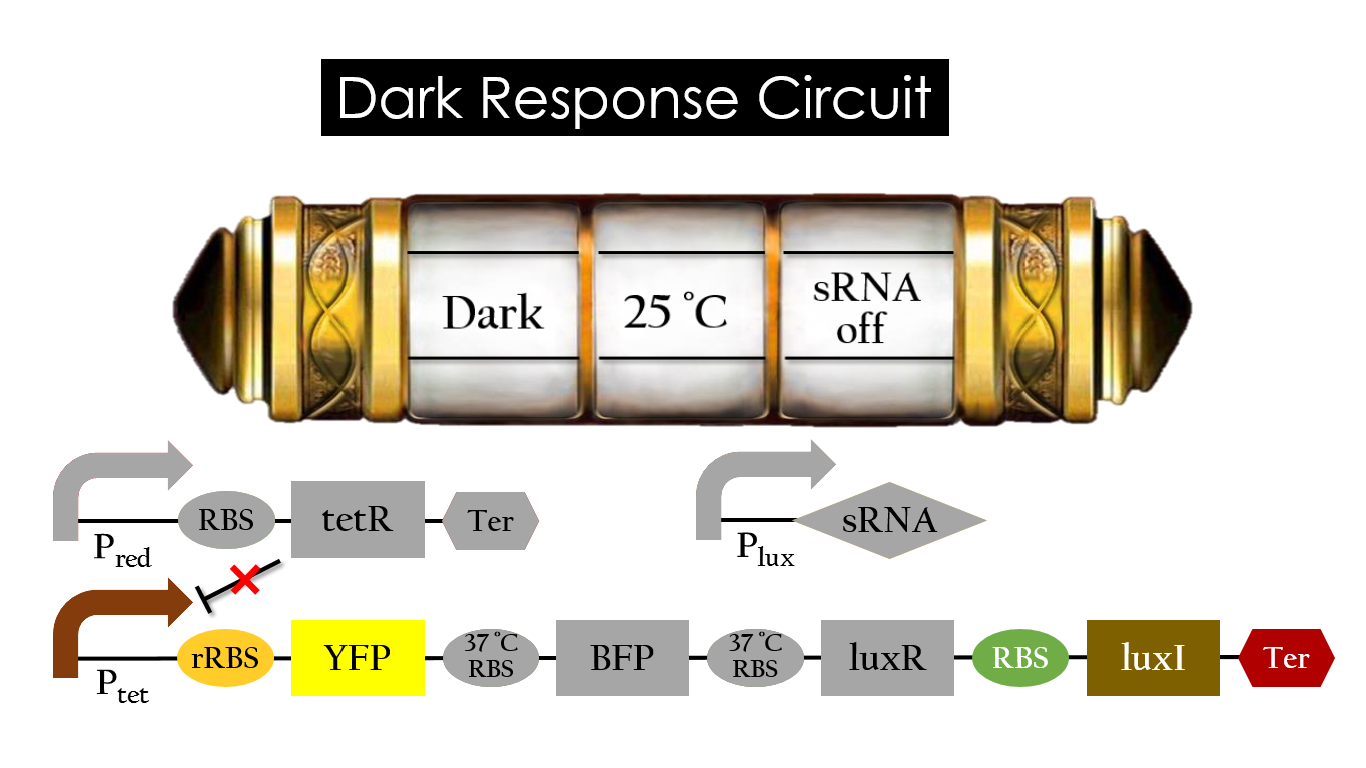
Pred activates in the presence of red light, producing tetR that represses Ptet. In other words, the dark induced part is inactive in the exposure of red light and active without red light. At 30°C in the dark, only normal RBS functions to express YFP while 37°C RBS remains as a hairpin structure.
At 37°C, however, 37°C RBS unfolds to express LuxR and BFP. Just like the mechanism employed in the light induced part, LuxR forms a complex with AHL to activate Plux that produces the sRNA to block normal RBS. This way, the YFP downstream of YFP cannot be expressed and BFP is expressed at 37°C
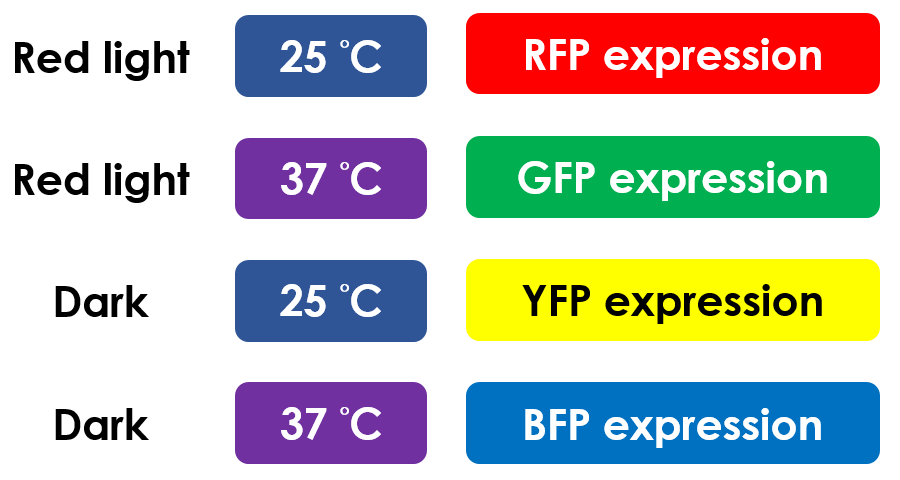
Pathway Regulation Summary
This is the pathway summary of the E.colightuner. There is one kind of fluorescent protein in each condition we set.
Future Work
The multiple regulated-system we have created in this project consisted of two different parts: light induced part and dark induced part. Each part regulates two different genes, resulting in a total of four genes that can be regulated. By adding new parts to the system, it is plausible that the system can regulate more genes. We tend to do that by employing more light sensing promoters. Each light sensor we add, we would be able to regulate two more different genes by employing regulation mechanism of 37°C RBS and sRNA.
On the other hand, we tend to optimize our system to achieve subtle control of the four gene expressions. We might be able to do this by taking the values between the limits we have set. Instead of 30°C and 37°C , we tend to take the values between so we can create a lot more conditions under which different level of expression can be achieved. We can also vary the intensity of red light to control the level of expression. The ultimate goal is to precisely control the level of expression of each gene.
Application
Taiwan is renowned for Phalaenopsis, producing more than 35 million Phalaenopsis each year. There is no doubt that Taiwan has become "The Kingdom of Phalaenopsis". The technique we usually use to grow Phalaenopsis is plant tissue culture, thus, we intend to apply our system to plant tissue culture as well.
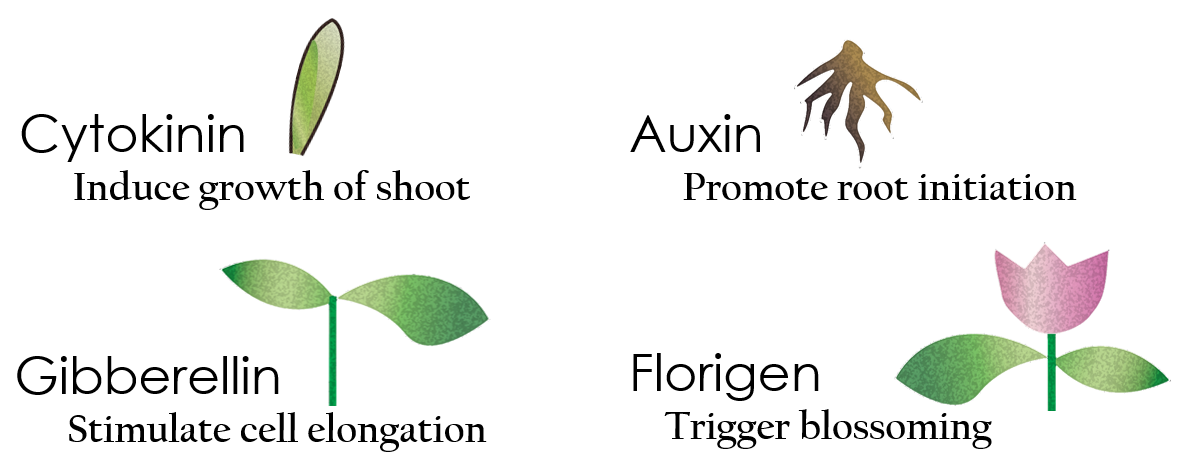
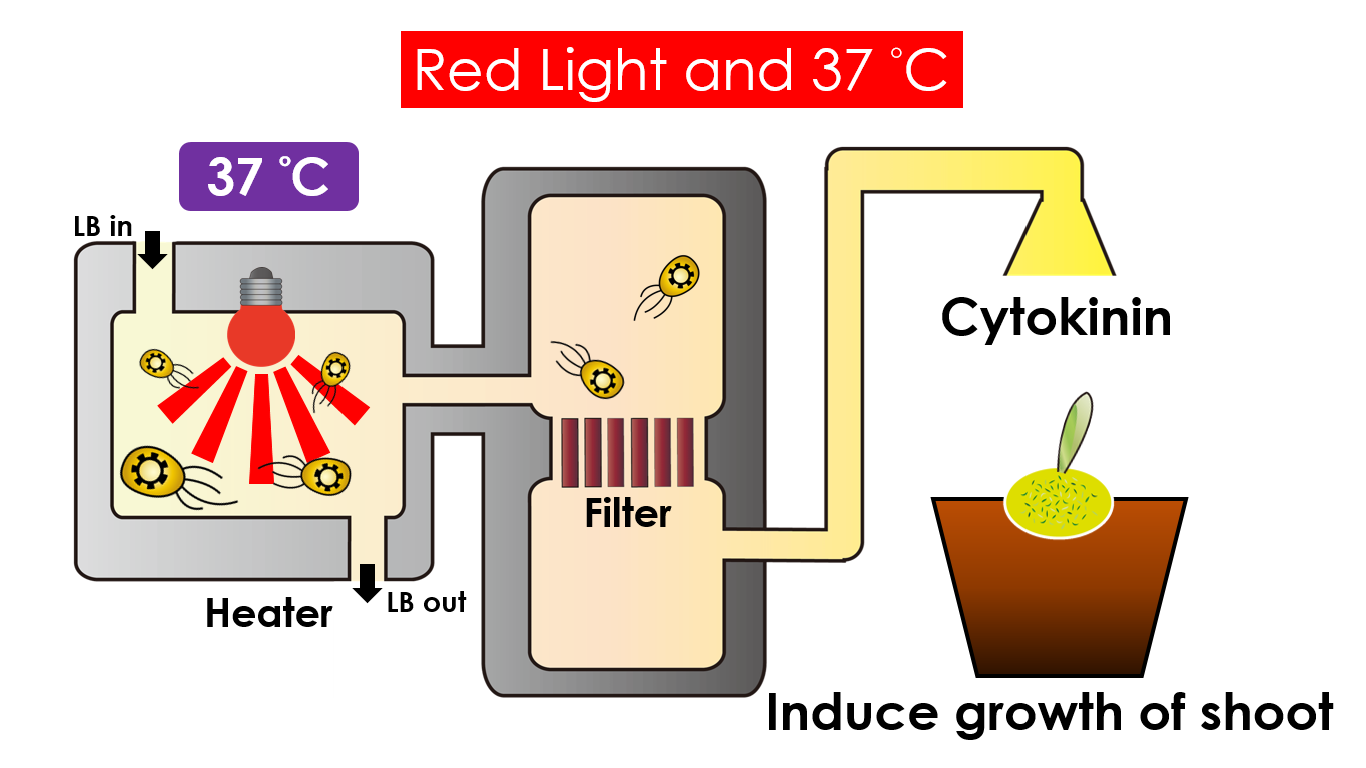
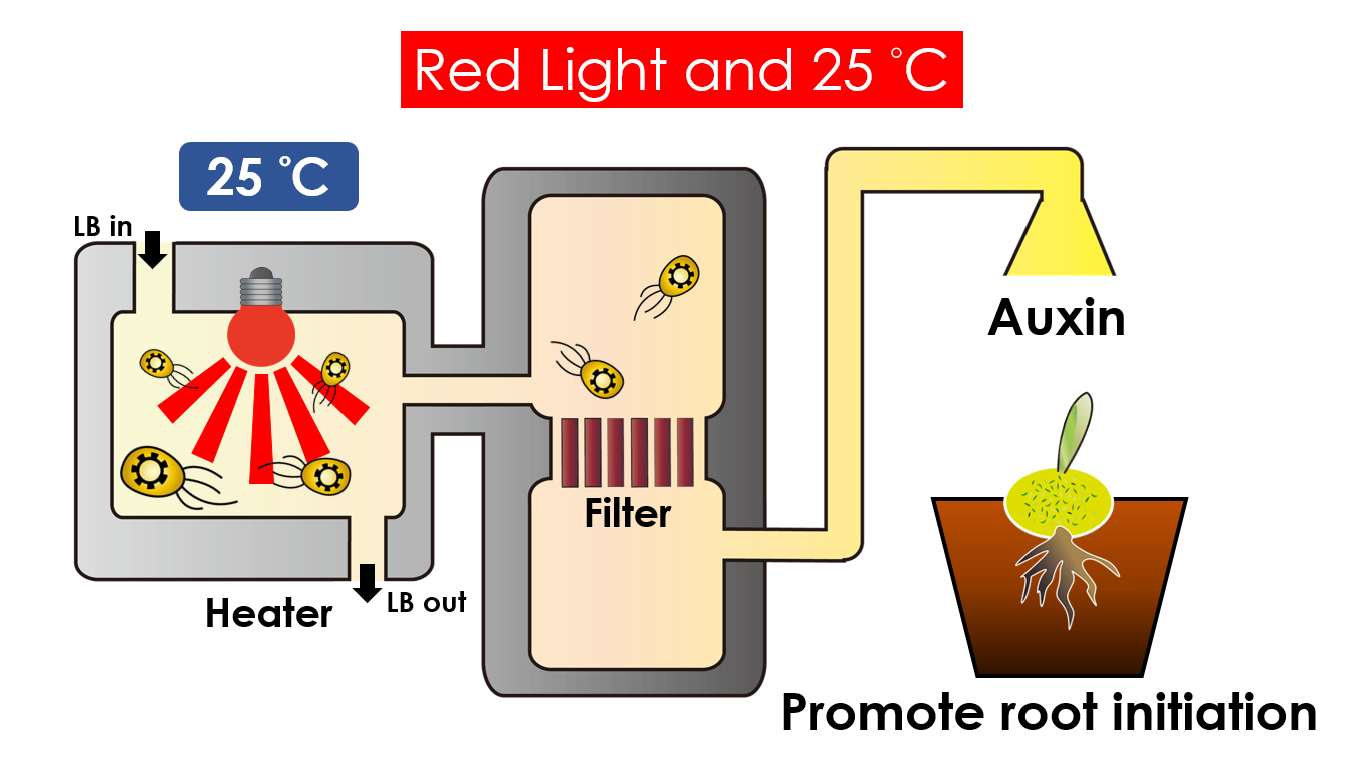
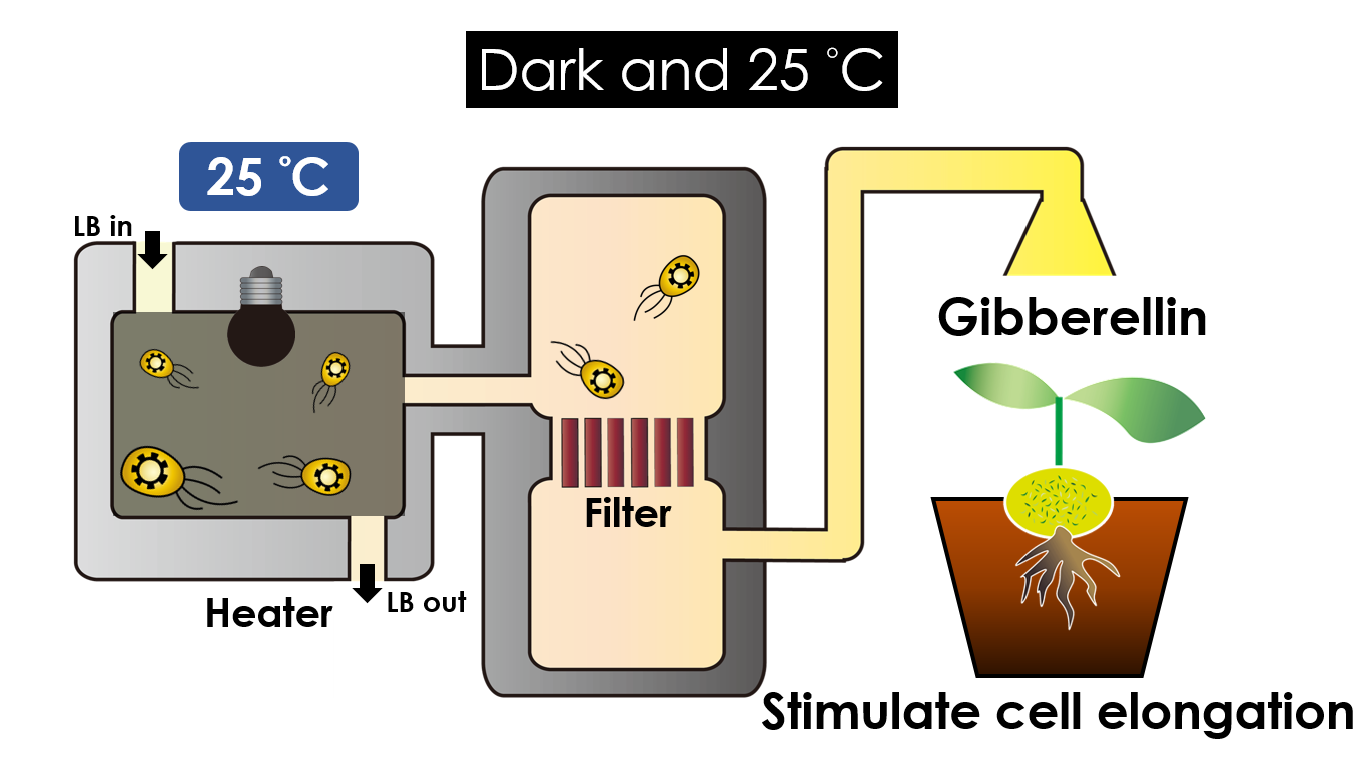
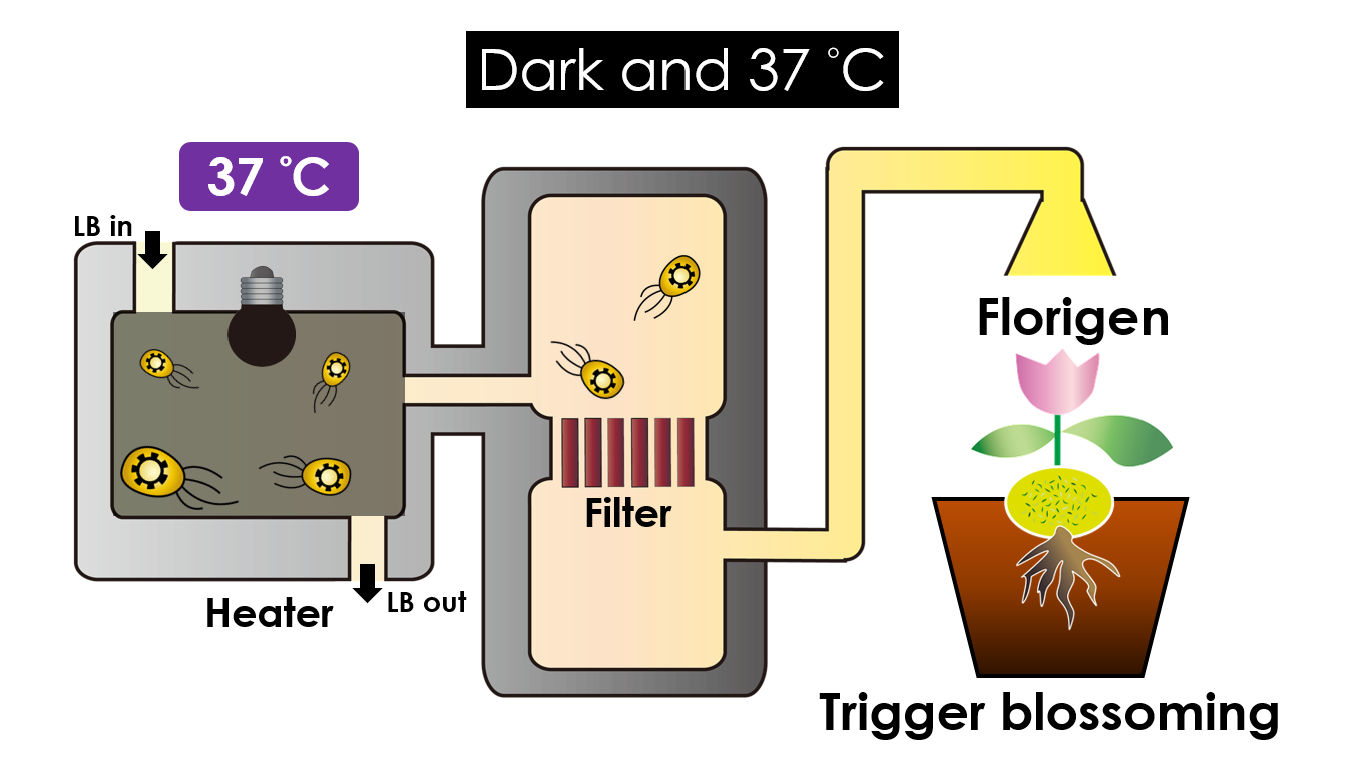
Tissue culture is the growth of tissues or cells separate from the organism. This is typically facilitated via use of a liquid, semi-solid, or solid growth medium, such as broth or agar. Moreover, hormones are added to the growth medium with a view to meeting what the plants need during growth period. However, the hormones needed during each growing state are not the same. So, we need to figure out a device which can generate multiple hormones. By adding our engineered E.coli to the growth medium, the plants can get multiple hormones secreted by E.coli under defferent circumstances. We chose four common hormones as our genes, Auxin, Cytokinin, Gibberellin, and Florigen.
Under red light and 37°C, Cytokinin will be produced. Cytokinin is a plant growth substance, which can primarily induce cell growth and differentiation in plant roots and shoots. It can decreace apical dominance, which will make the main stem to grow better than other stem. So the stem will all become stronger. Cytokinin can also signal lateral bud growth, which will make the plant bushier.
Under red light and 30°C, Auxin will be produced. Auxin can coordinate development from cellular, organs, to whole plant. Auxin molecules in cells may cause direct response by stimulation, inhibition of certain gene’s expression. On the cellular level, Auxins is important for cell growth. It can induced axial elongation in roots and lateral expansion which can make the roots swelling. For the whole plant level, it affect the shape of the plant. Because of different function, each organ have uneven Auxin and cause the specific shape.
Under dark and 30°C, Gibberellin will be produced. Gibberellin is important in elongation of cell. It cause the extension of stem by inducing cell division and elongation. A special characteristic of it is that it can produce more mass when the plant is exposed under cold temperature.
Under dark and 37°C, Florigen will be produced. Florigen is the key to trigger the flower. The signal of flower is first start by the production of mRNA coding CO( a transcription factor). CO mRNA then translate into CO protein. CO protein then promote transcription of FT gene, and FT gene translate in FT protein. FT protein will be transported via the phloem to shoot apical meristem, where FT protein will interact with FD protein(transcription factor) to activate floral identity, thus inducing flowering.
 "
"