Header
Contents |
Overview
How can one bind Nanoparticles to bacteria?
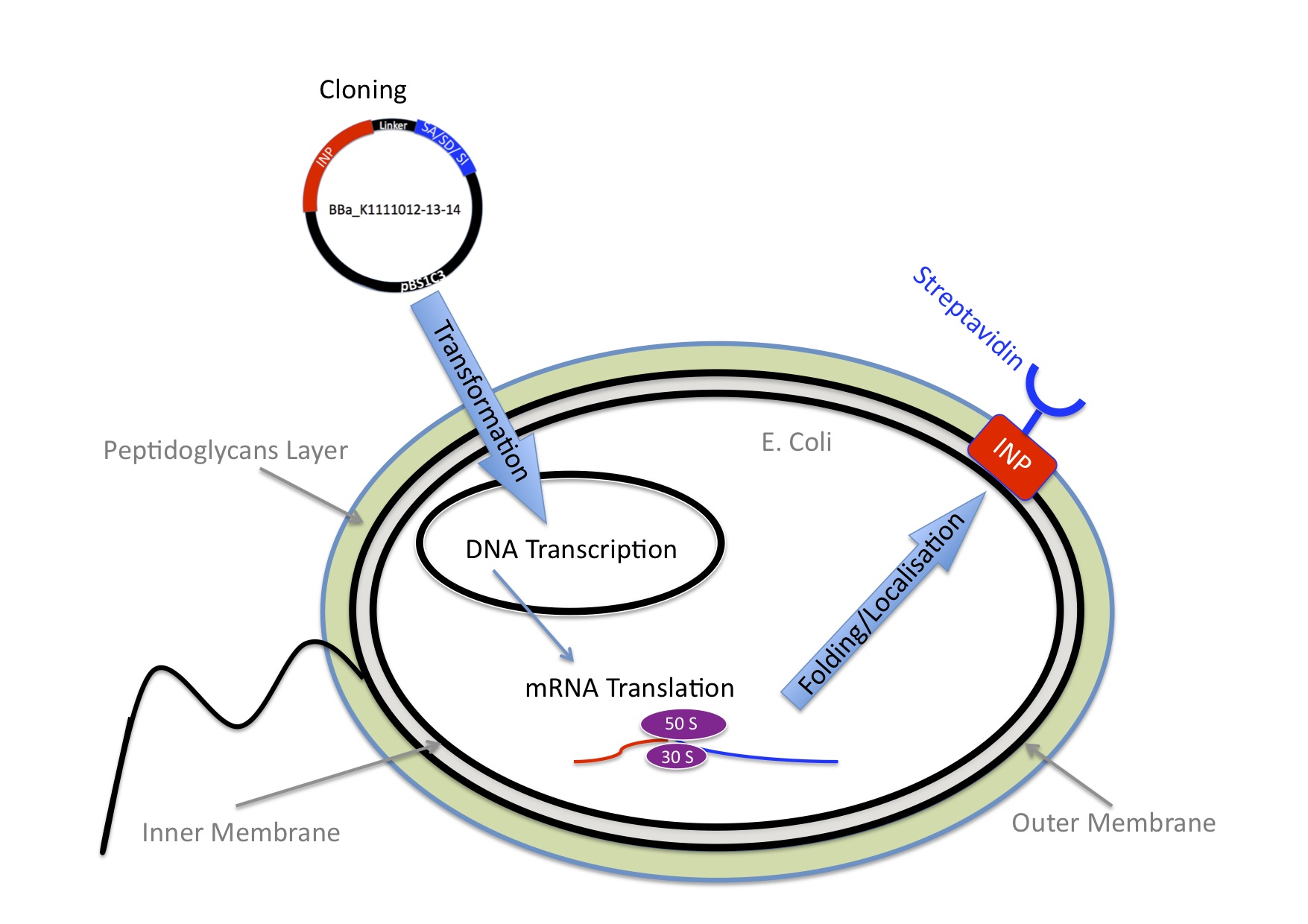
Once we knew we wanted to bind Nanoparticles to bacteria we needed an idea how to connect those two components. We came up with the idea to use one of the strongest non covalent interactions existing in nature, the interaction between Streptavidin and Biotin with a Kd of 10-14 mol/l. Streptavidin is a tetrameric protein, each of the four subunits can bind one biotin. The Streptavidin-Biotin complex is resistant against different pHs, temperatures and detergents. Also streptavidin binds only rarely unspecific. The bacteria need to express Streptavidin on its surface and the Nanoparticles needed to be biotynilated.
INP-Streptavidin
E.Coli are Gram-negative bacteria and they have two membranes, an inner one and an outer one, separated by a periplasmic space. Furthermore, the outer membrane is covered with a glycocalix, a layer of sugars. (add image) There are different strategies to express a heterologous protein on the outer membrane of E.coli. Peptides can be inserted in outer membrane proteins, or on fimbriae, fused to beta-autotransporters or lipoproteins, as for example the (Lpp)-OmpA protein5 which was used by the igem team 2006 of Harvard and the igem team 2009 of Washington. But then there is also another promising display system that uses ice nucleation protein (INP) from pseusomonas syringae, which is a glycosyl-phosphatidyliositol anchored outer membrane protein. The GPI anchor is normally used by eukaryotes, so to see this motif in prokaryotes is quite unique.7 Over 90% of INP is found in the outer membrane when it is expressed in E.coli (Wolber et al). It has successfully been used for cell surface display of enzymes, single-chain antibodies and antigens.9 Jung et al (1998) successfully expressed functional levansucrase on the surface of E.coli by fusing it to INP, levansucrase is a 47 kDA protein. Streptavidin is only 14 kDa, so we assumed that it should be even easier for the E.coli to export it.
Streptavidin originates from the bacterium Streptomyces avidinii. It is a tetrameric protein, which means it has 4 subunits, each of them can bind a biotin molecule. For our Biobricks we used different kinds of streptavidin. The Biobrick BBa_K1111014 contains streptavidin that was cloned out of the Biobrick BBa_K283010.
The Biobrick BBa_K1111012 has a monomeric streptavidin in it, which means only one of the four subunits of streptavidin is able to bind to biotin. This decreases the dissociation rate. The plasmid containing the sequence was provided by the laboratory of Mark Howarth at Oxford university Dept. of Biochemistry (http://users.ox.ac.uk/~bioc0756/MyWebs/activesite/index.htm). The Biobrick BBa_K1111013 was designed to serve as a negative control for our experiments, it contains a dead version of streptavidin, which means it doesn’t bind to biotin.
Our Biobricks
INP-YFP
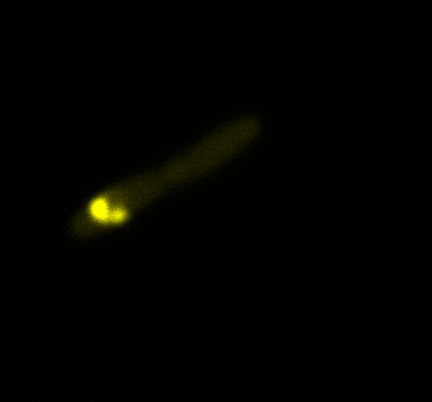
There was already a Biobrick that contained INP in the registry (BBa_K523013), so we decided to use it. It was designed by the Edinburgh igem team of 2011 (https://2011.igem.org/Team:Edinburgh/Cell_Display). This Biobrick encodes a fusion protein of a truncated and codon optimized INP and yellow fluorescent protein (YFP) under control of the LacZ promoter. The INP is truncated, it only contains the first 211 and last 97 amino acids that are normally encoded by the inaK gene. To characterize the Biobrick BBa_K523013 we used a confocal microscope to see if it was expressed.
Looking at the image we realized that there were some points that showed very strong fluorescence ,
that the team of Edinburgh didn’t describe. We continued characterizing this biobrick by first determing the percentage of bacteria that was fluorescent, approximatively 30% of E.coli expressed YFP. We also counted the percentage of fluorescent bacteria that showed these points of very strong fluorescence, around XX%. We started looking for an explanation for this phenomena and found in the literature1, 2 that INP forms aggregates in the cell membrane, to have good ice nucleation abilities. So it suggests, that this also happens if INP is fused to another protein.
The team of Edinburgh used an image of eppendorf tube to proof surface localization, because their imaging technology wasn’t good enough. So we looked at it with a confocal microscope (image), but we too weren’t able to show surface localization by simply looking at the bacteria. So we performed an immunofluorescent assay with an anti YFP antibody. Then we made images with the microscope first a bright field, then exiting the YFP, and finally exiting the Antibody (red) By superimposing the images we could show nicely that the YFP is located at the outer membrane of the E.coli (image)
Results
In the following section, you will find results concerning our cell surface display work. Before moving on it is important to define the nomenclature we will further use: 1)Initially used plasmid: INP_EYFP Biobrick: <address><a href="http://parts.igem.org/Part:BBa_K523013">BBa_K523013</a></address> Streptavidin Alive: <a href="http://parts.igem.org/Part:BBa_K1111010">BBa_K1111010</a> Streptavidin Dead: <a href="http://parts.igem.org/Part:BBa_K1111011">BBa_K1111011</a> Streptavidin iGEM: <a href="http://parts.igem.org/Part:BBa_K283010">BBa_K283010</a>
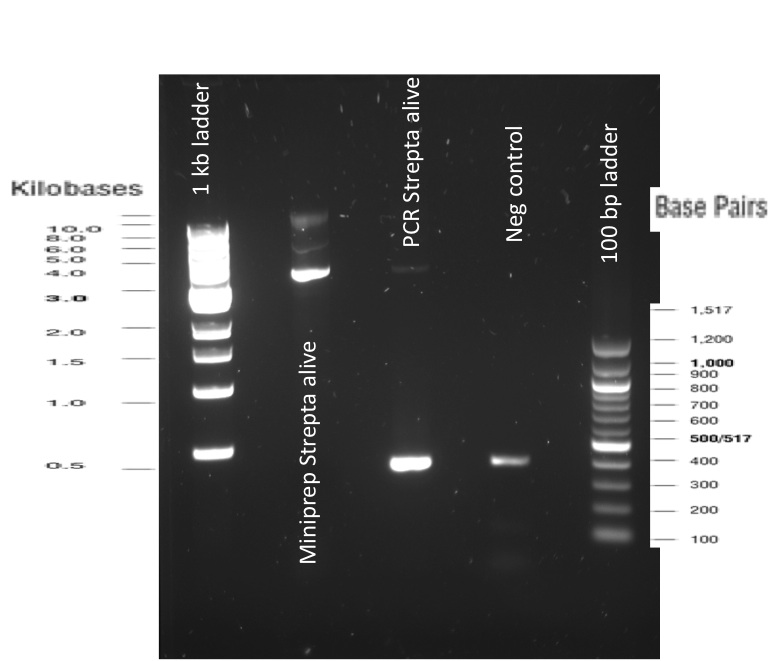
2) Gibson reaction product names: INP_construct = BBa_K523013 without EYFP ISA = INP_Streptavidin Alive gibson product <a href="http://parts.igem.org/Part:BBa_K1111012">BBa_K1111012</a> ISD = INP_Streptavidin Dead gibson product<a href="http://parts.igem.org/Part:BBa_K1111013">BBa_K1111013</a> ISI = INP_Streptavidin iGEM BBa_K283010 gibson product<a href="http://parts.igem.org/Part:BBa_K1111014">BBa_K1111014</a> 2) PCR reactions products SA = Streptavidin Alive, SD = Streptavidin Dead, SI = Streptavidin iGEM
Indeed to trace back what we did, the first steps were perform Polymerase Chain Reactions (PCR) in order to isolate the our Bricks.
References
 "
"