Team:TU-Munich/Notebook/Labjournal
From 2013.igem.org

Labjournal
Week 1
Monday, April 22nd
Transformation of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Transformation of Phytochrome B for protein fusion.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Sequencing of RFP-Generator (RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Sequencing of RFP-Generator (RFC25, pSB1C3)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
Tuesday, April 23rd
Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Procedure:
- pSB1C3 plasmid with BBa_K801031 (PhyB 2 - 908 aa, RFC25): Colonies were picked from chloramphenicol plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 4 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
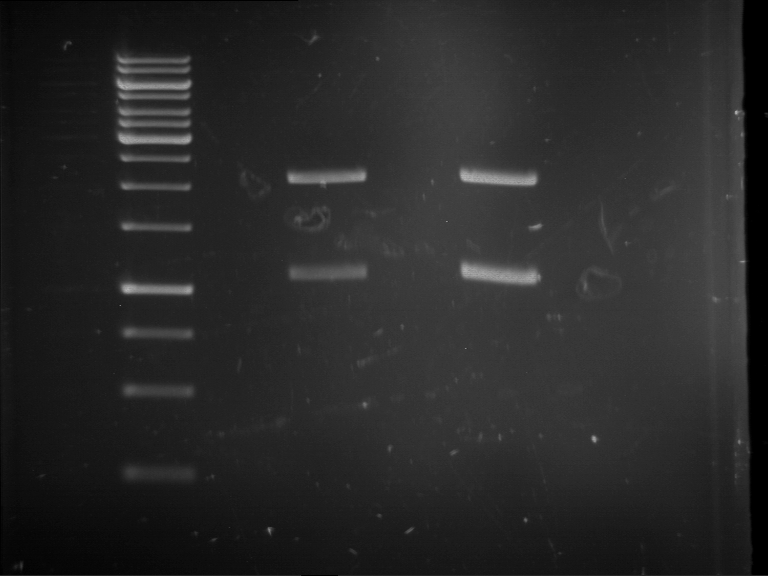
Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5)
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5).
Procedure:
- Batch for analytical digestion for P4 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P4 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P5 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P5 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P4 | P5 |
| Mutation successful | Mutation successful! |
- Parts are compliant and do not contain RFC25 forbidden restriction sites.
Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different vectors we sequenced received the following barcodes:
- ADH in pTUM100: FR01002265
- TEF1 in pTUM100: FR01002266
- TEF2 in pTUM100: FR01002266
- GAL in pTUM100: FR01002268
Sequencing of TEF2 in pTUM100 was not interpretable. The other sequences were consistent with the sequences in the parts registry.
Wednesday, April 24th
Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
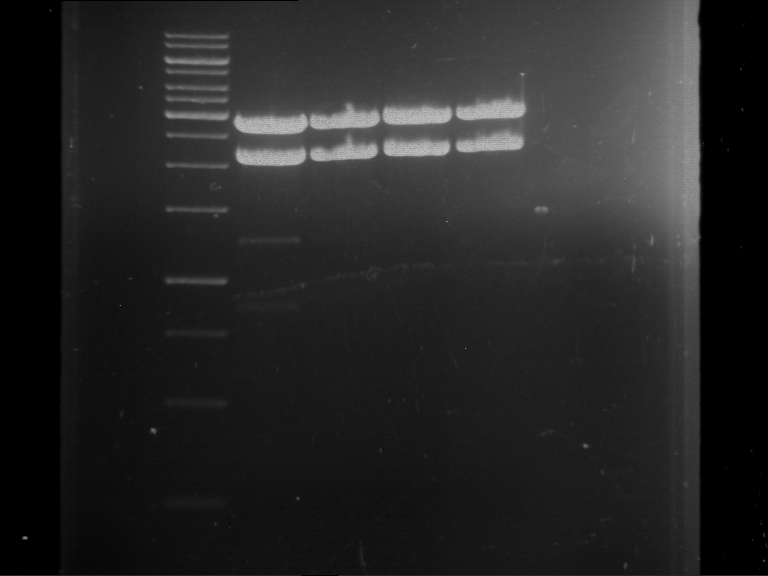
Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10.
Procedure:
- Batch for analytical digestion for P7 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P7 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P8 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P8 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P9 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P9 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P10 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P10 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | P7 | P8 | P9 | P10 |
| Part is correct | Part is correct | Part is correct | Part is correct |
Transformation of E. coli XL1 blue with EreA and EreB (pDEST14)
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Transformation of E. coli XL1 blue with EreA and EreB (pDEST14).
Procedure:
- Plasmid DNA was received dried in paper from McMaster University.
- DNA was resuspended in ddH2O
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
Week 2
Monday, April 29nd
Miniprep of pRK792, pRK793, pRIL, ereA, ereB
Investigator: Leonie
Aim of the experiment: Miniprep of pRK792, pRK793, pRIL, ereA, ereB
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations were determined by measuring the absorption:
| Plasmid | c [ng/µl] |
| pRK792 | 214,5 |
| pRK793 | 154,3 |
| pRIL | 6,1 |
| ereA | 183,4 |
| ereB | 98,3 |
- The procedure will have to be repeated for pRIL
Midiprep of RFC25 compatible RFP generator in (pSB1C3)
Investigator: Andreas, Florian
Aim of the experiment: Midiprep of RFC25 compatible RFP generator in (pSB1C3)
Procedure:
- Midiprep was performed after manufacturer's protocol (QIAprep Midiprep, QIAGEN)
Tuesday, April 30th
Preparative digestion and gelelectrophoresis of mINF1 and Leptin with HindIII and EheI
Investigator: Louise, Johanna
Aim of the experiment: Testing the enzymes, since the earlier digestion with probably older enzymes did not work (no bands on gel) .
Procedure:
- Batch for preparative digestion for mINF1 with HindIII and EheI
| volume | reagent |
| 10 µl | Plasmid DNA mINF11 |
| 5 µl | Tango Buffer (10x) |
| 2 µl | HindIII (10 U/µl) |
| 3 µl | EheI (10 U/µl) |
| 30 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion for Leptin with HindIII and EheI
| volume | reagent |
| 20 µl | Plasmid DNA Leptin |
| 5 µl | Tango Buffer (10x) |
| 2 µl | HindIII (10 U/µl) |
| 3 µl | EheI (10 U/µl) |
| 20 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C.
Transformation of E. coli XL1 blue with PSH21 (P17)
Investigator: Johanna, Louise
Aim of the experiment: Transformation of E. coli XL1 blue with PSH21.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice for 10 minutes.
- 5 µl of DNA was added to 250 µl of competent cells and gently mixed.
- 60 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding the DNA to 2 ml LB-medium.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
Friday, May 3th
Transformation of E. coli XL1 blue with pSB1C3-GFP, pSB1C3-mkate2, pSB1C3-mCherry
Investigator: Andreas
Aim of the experiment: Transformation of E. coli XL1 blue with pSB1C3-GFP, pSB1C3-mkate2, pSB1C3-mCherry.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice for 10 minutes.
- 5 µl of DNA was added to 250 µl of competent cells and gently mixed.
- 60 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding the DNA to 2 ml LB-medium.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
Plating of received E. coli containing biobricks
Investigator: Jeff
Aim of the experiment: The received biobricks were already transformed in E. coli and were in an agar stabs. These E. coli cells were transferred with an inoculation loop on antibiotic selection plates and were incubated over night.
Operational sequence:
- Bacterias containing plasmids with biobricks were transferred with a sterile inoculation loop on antibiotic plates and were incubated at 37 °C overnight.
- The biobricks were:
| Name | Function |
|---|---|
| [[http://partsregistry.org/Part:BBa_K157004 BBa_K157004]] | Fluoresceine A -binding derivative of a Lipocalin |
| [[http://partsregistry.org/Part:BBa_K863000 BBa_K863000]] | bpul (laccase from Bacillus pumilus) with T7 promoter, RBS and HIS tag |
| [[http://partsregistry.org/Part:BBa_K909009 BBa_K909009]] | cDNA of UV-B sensing protein UVR8 from Arabidopsis thaliana |
| [[http://partsregistry.org/Part:BBa_K337013 BBa_K337013]] | constitutive promoter SV40 |
| [[http://partsregistry.org/Part:BBa_K747096 BBa_K747096]] | constitutive promoter CMV |
| [[http://partsregistry.org/Part:BBa_K414002 BBa_K414002]] | constitutive promoter CaMV35S |
| [[http://partsregistry.org/Part:BBa_K147002 BBa_K147002]] | xylE gene coding for catechol-1,2-dioxygenase |
| [[http://partsregistry.org/Part:BBa_E2020 BBa_E2020]] | Cerulean |
| [[http://partsregistry.org/Part:BBa_K404319 BBa_K404319]] | mCFP |
| [[http://partsregistry.org/Part:BBa_E0040 BBa_E0040]] | GFPmut3b |
| [[http://partsregistry.org/Part:BBa_K592010 BBa_K592010]] | amilGFP |
| [[http://partsregistry.org/Part:BBa_E2050 BBa_E2050]] | mOrange |
| [[http://partsregistry.org/Part:BBa_K399001 BBa_K399001]] | mStrawberry |
| [[http://partsregistry.org/Part:BBa_E1010 BBa_E1010]] | mRFP1 |
| [[http://partsregistry.org/Part:BBa_E2060 BBa_E2060]] | mCherry |
| [[http://partsregistry.org/Part:BBa_K592012 BBa_K592012]] | eforRed |
| [[http://partsregistry.org/Part:BBa_K592011 BBa_K592011]] | cjBlue |
| [[http://partsregistry.org/Part:BBa_K864401 BBa_K864401]] | aeBlue |
| [[http://partsregistry.org/Part:BBa_K592009 BBa_K592009]] | amilCP, blue chromoprotein |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414000 BBa_K414000]] | RD29A Promoter + Strong Plant Kozak (RBS) + RFP |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414006 BBa_K414006]] | 35S Promoter + (Strong Plant RBS + GFP) From K414001 |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414007 BBa_K414007]] | DREB1C promoter |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K678001 BBa_K678001]] | alcA promoter |
| [[http://partsregistry.org/Part:BBa_K243004 BBa_K243004]] | Short linker |
| [[http://partsregistry.org/Part:BBa_K243005 BBa_K243005]] | Middle linker |
| [[http://partsregistry.org/Part:BBa_K243006 BBa_K243006]] | Long Linker |
| [[http://partsregistry.org/Part:BBa_K243029 BBa_K243029]] | GSAT Linker |
| [[http://partsregistry.org/Part:BBa_K243030 BBa_K243030]] | SEG Linker
|
 "
"