Team:TU-Munich/Notebook/Labjournal
From 2013.igem.org

Labjournal
Week 1
Monday, April 22nd
Transformation of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Transformation of Phytochrome B for protein fusion.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Miniprep of pTUM100 with pGAL, pTEF1, pTEF2, pADH and RFC25 compatible RFP generator
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Sequencing of RFP-Generator (RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Sequencing of RFP-Generator (RFC25, pSB1C3)
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer)
Tuesday, April 23rd
Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Picking of of E. coli XL1 blue with Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Procedure:
- pSB1C3 plasmid with BBa_K801031 (PhyB 2 - 908 aa, RFC25): Colonies were picked from chloramphenicol plates.
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 4 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight
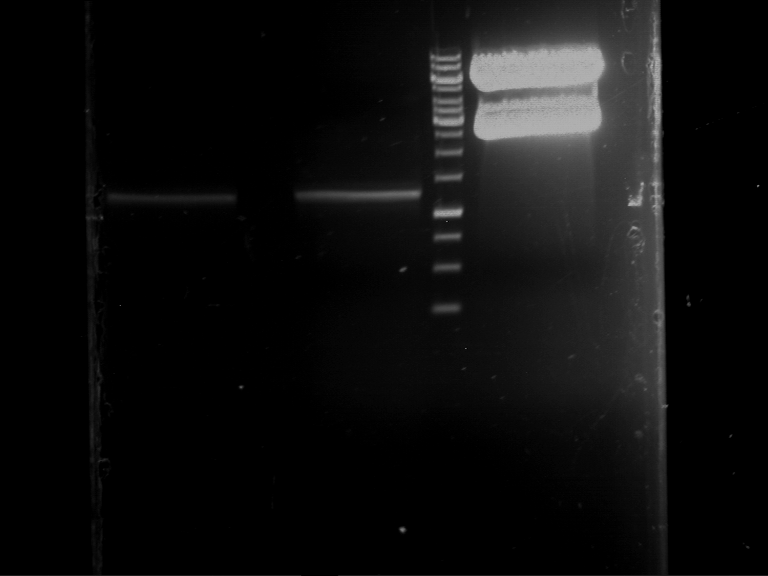
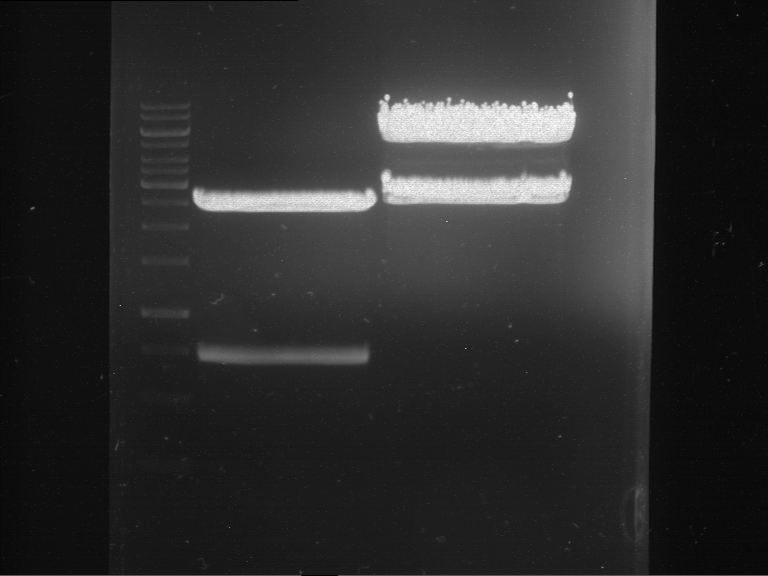
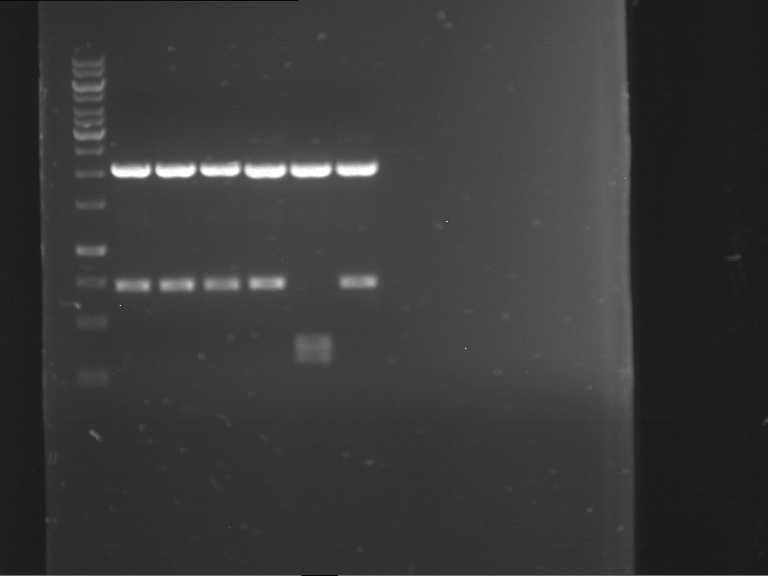
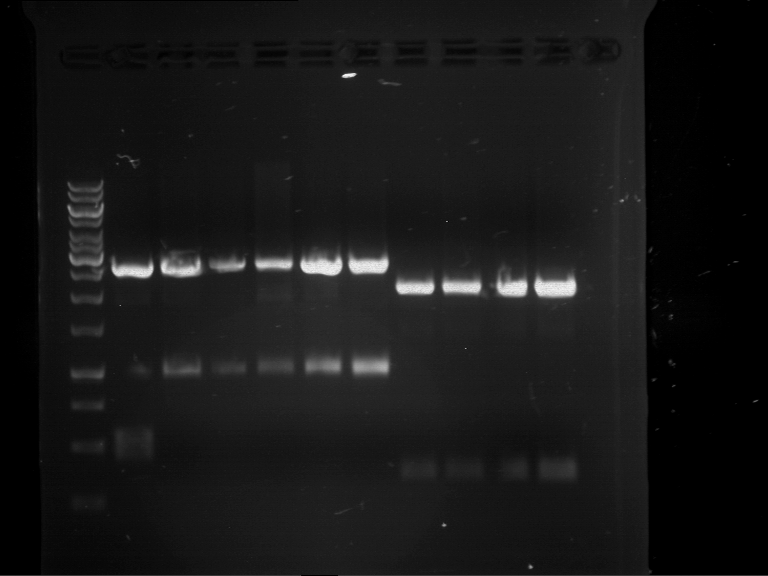
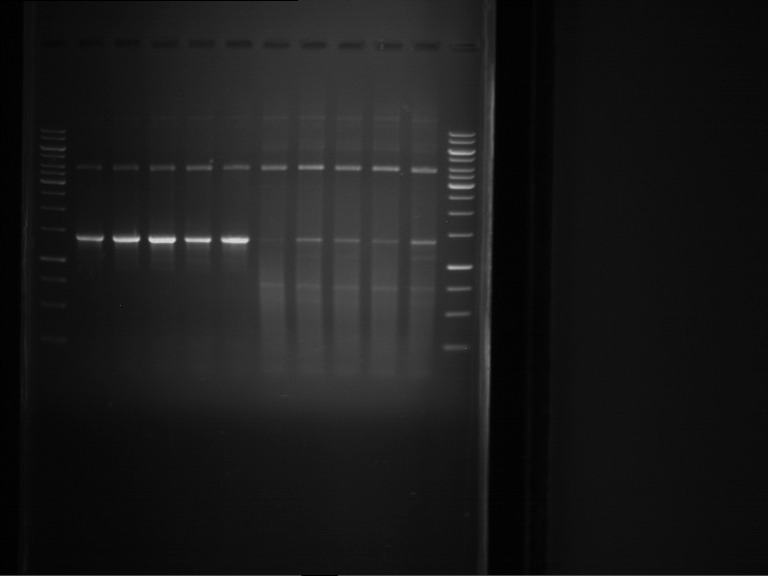
Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5)
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of RFP-generator (RFC25, pSB1C3, P4 & P5).
Procedure:
- Batch for analytical digestion for P4 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P4 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P5 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P5 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
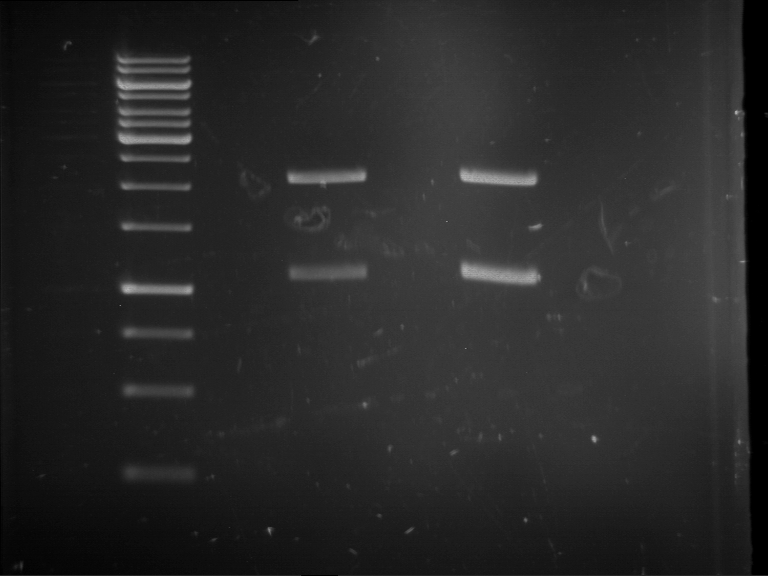
Results:
| 1 kbp ladder DNA ladder | P4 | P5 |
| Mutation successful | Mutation successful! |
- Parts are compliant and do not contain RFC25 forbidden restriction sites.
Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Investigator: Jeff, Leonie, Rosario, Florian
Aim of the experiment: Sequencing of pTUM vectors with pGAL, pADH, pTEF1, pTEF2
Procedure:
Sequencing batch were prepared after manufacturer's protocol. (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different vectors we sequenced received the following barcodes:
- ADH in pTUM100: FR01002265
- TEF1 in pTUM100: FR01002266
- TEF2 in pTUM100: FR01002266
- GAL in pTUM100: FR01002268
Sequencing of TEF2 in pTUM100 was not interpretable. The other sequences were consistent with the sequences in the parts registry.
Wednesday, April 24th
Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3)
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Miniprep of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
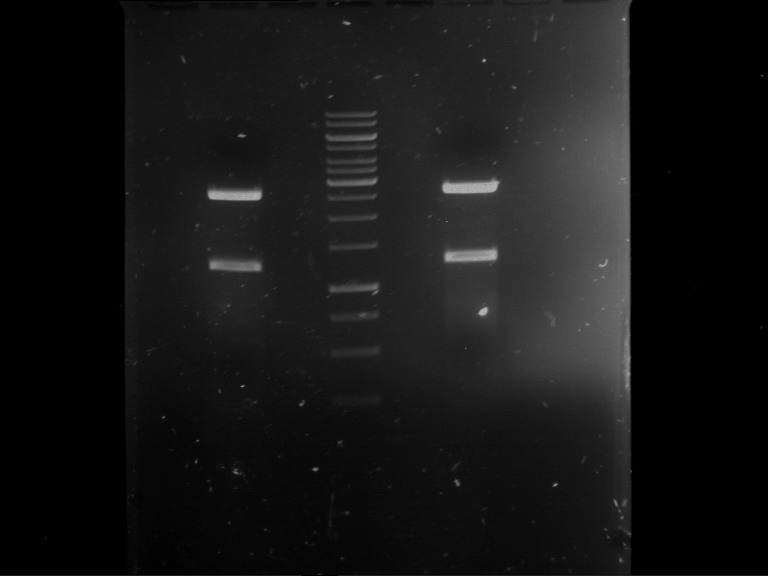
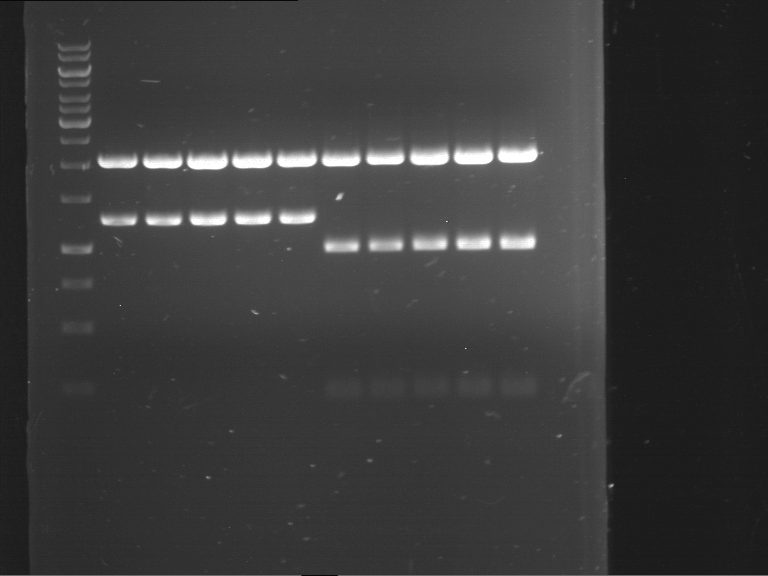
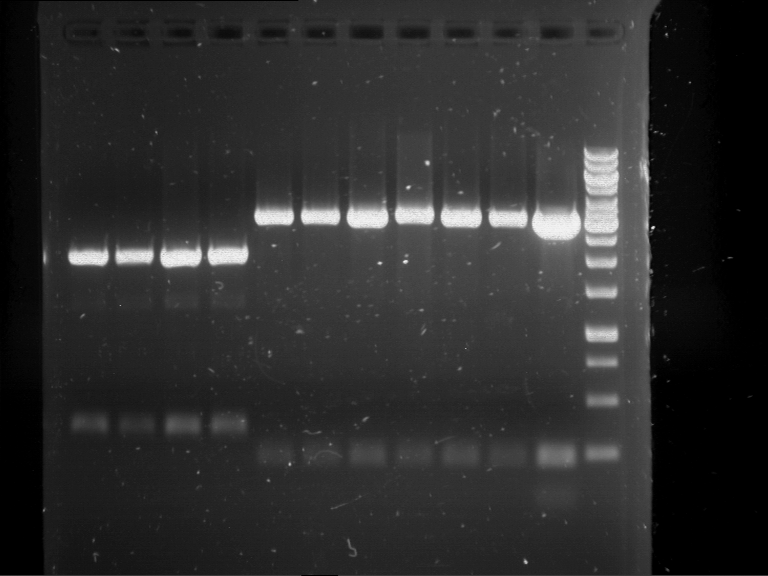
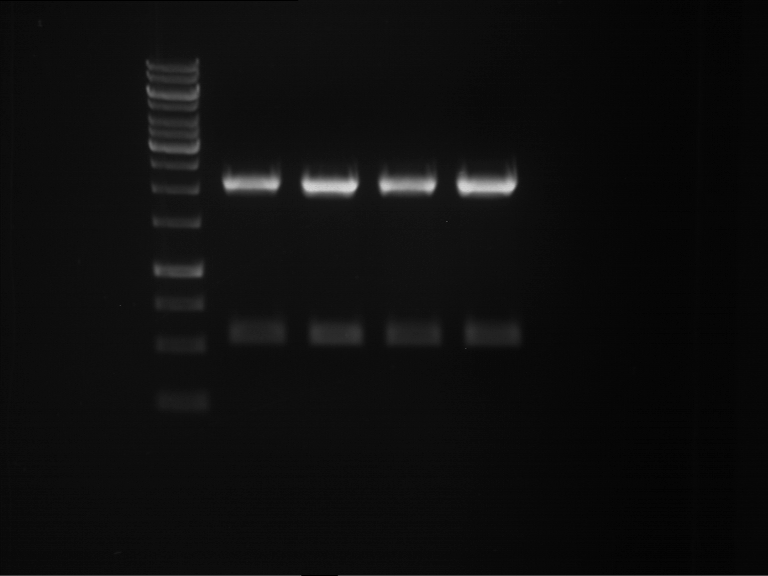
Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Analytical digestion and gelelectrophoresis of Phytochrome B (2-908 N-terminal amino acids) (BBa_K801031, RFC25, pSB1C3), P7 - P10.
Procedure:
- Batch for analytical digestion for P7 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P7 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P8 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P8 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P9 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P9 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Batch for analytical digestion for P10 with NgoMIV+AgeI-HF
| volume | reagent |
| 2.5 µl | Plasmid DNA P10 |
| 2 µl | NEBuffer 4 (10x) |
| 0.25 µl | NgoMIV (10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
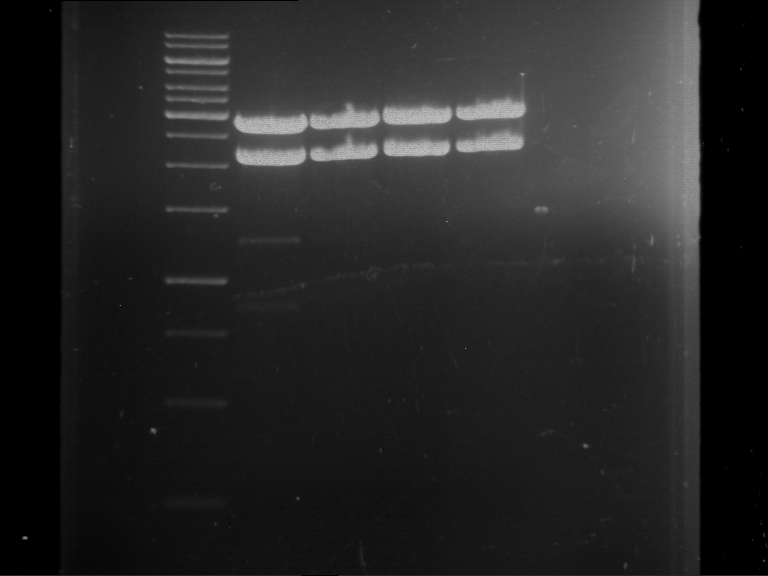
Results:
| 1 kbp ladder DNA ladder | P7 | P8 | P9 | P10 |
| Part is correct | Part is correct | Part is correct | Part is correct |
Transformation of E. coli XL1 blue with EreA and EreB (pDEST14)
Investigator: Jeff, Leonie, Florian
Aim of the experiment: Transformation of E. coli XL1 blue with EreA and EreB (pDEST14).
Procedure:
- Plasmid DNA was received dried in paper from McMaster University.
- DNA was resuspended in ddH2O
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
Week 2
Monday, April 29nd
Miniprep of pRK792, pRK793, pRIL, ereA, ereB
Investigator: Leonie
Aim of the experiment: Miniprep of pRK792, pRK793, pRIL, ereA, ereB
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Concentrations were determined by measuring the absorption:
| Plasmid | c [ng/µl] |
| pRK792 | 214,5 |
| pRK793 | 154,3 |
| pRIL | 6,1 |
| ereA | 183,4 |
| ereB | 98,3 |
- The procedure will have to be repeated for pRIL
Midiprep of RFC25 compatible RFP generator in (pSB1C3)
Investigator: Andreas, Florian
Aim of the experiment: Midiprep of RFC25 compatible RFP generator in (pSB1C3)
Procedure:
- Midiprep was performed after manufacturer's protocol (QIAprep Midiprep, QIAGEN)
Tuesday, April 30th
Preparative digestion and gelelectrophoresis of mINF1 and Leptin with HindIII and EheI
Investigator: Louise, Johanna
Aim of the experiment: Testing the enzymes, since the earlier digestion with probably older enzymes did not work (no bands on gel) .
Procedure:
- Batch for preparative digestion for mINF1 with HindIII and EheI
| volume | reagent |
| 10 µl | Plasmid DNA mINF11 |
| 5 µl | Tango Buffer (10x) |
| 2 µl | HindIII (10 U/µl) |
| 3 µl | EheI (10 U/µl) |
| 30 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion for Leptin with HindIII and EheI
| volume | reagent |
| 20 µl | Plasmid DNA Leptin |
| 5 µl | Tango Buffer (10x) |
| 2 µl | HindIII (10 U/µl) |
| 3 µl | EheI (10 U/µl) |
| 20 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C.
Transformation of E. coli XL1 blue with PSH21 (P17)
Investigator: Johanna, Louise
Aim of the experiment: Transformation of E. coli XL1 blue with PSH21.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice for 10 minutes.
- 5 µl of DNA was added to 250 µl of competent cells and gently mixed.
- 60 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding the DNA to 2 ml LB-medium.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
Thursday, May 2nd
Miniprep of pSH21 (pAutoRex8) containing AlcR/AlcA promotor system
Investigator: Katrin
Aim of the experiment: Miniprep of pSH21 (pAutoRex8) containing AlcR/AlcA promotor system
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The concentration was measured (900 ng/µl)
Friday, May 3rd
Transformation of E. coli XL1 blue with pSB1C3-GFP, pSB1C3-mkate2, pSB1C3-mCherry
Investigator: Andreas
Aim of the experiment: Transformation of E. coli XL1 blue with pSB1C3-GFP, pSB1C3-mkate2, pSB1C3-mCherry.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice for 10 minutes.
- 5 µl of DNA was added to 250 µl of competent cells and gently mixed.
- 60 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding the DNA to 2 ml LB-medium.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- The transformated cells were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated on chlorampenicol and ampicillin plates.
Plating of received E. coli containing biobricks
Investigator: Jeff, Andreas
Aim of the experiment: The received biobricks were already transformed in E. coli and were in an agar stabs. These E. coli cells were transferred with an inoculation loop on antibiotic selection plates and were incubated over night.
Operational sequence:
- Bacterias containing plasmids with biobricks were transferred with a sterile inoculation loop on antibiotic plates and were incubated at 37 °C overnight.
- The biobricks were:
| Name | Function |
|---|---|
| [[http://partsregistry.org/Part:BBa_K157004 BBa_K157004]] | Fluoresceine A -binding derivative of a Lipocalin |
| [[http://partsregistry.org/Part:BBa_K863000 BBa_K863000]] | bpul (laccase from Bacillus pumilus) with T7 promoter, RBS and HIS tag |
| [[http://partsregistry.org/Part:BBa_K909009 BBa_K909009]] | cDNA of UV-B sensing protein UVR8 from Arabidopsis thaliana |
| [[http://partsregistry.org/Part:BBa_K337013 BBa_K337013]] | constitutive promoter SV40 |
| [[http://partsregistry.org/Part:BBa_K747096 BBa_K747096]] | constitutive promoter CMV |
| [[http://partsregistry.org/Part:BBa_K414002 BBa_K414002]] | constitutive promoter CaMV35S |
| [[http://partsregistry.org/Part:BBa_K147002 BBa_K147002]] | xylE gene coding for catechol-1,2-dioxygenase |
| [[http://partsregistry.org/Part:BBa_E2020 BBa_E2020]] | Cerulean |
| [[http://partsregistry.org/Part:BBa_K404319 BBa_K404319]] | mCFP |
| [[http://partsregistry.org/Part:BBa_E0040 BBa_E0040]] | GFPmut3b |
| [[http://partsregistry.org/Part:BBa_K592010 BBa_K592010]] | amilGFP |
| [[http://partsregistry.org/Part:BBa_E2050 BBa_E2050]] | mOrange |
| [[http://partsregistry.org/Part:BBa_K399001 BBa_K399001]] | mStrawberry |
| [[http://partsregistry.org/Part:BBa_E1010 BBa_E1010]] | mRFP1 |
| [[http://partsregistry.org/Part:BBa_E2060 BBa_E2060]] | mCherry |
| [[http://partsregistry.org/Part:BBa_K592012 BBa_K592012]] | eforRed |
| [[http://partsregistry.org/Part:BBa_K592011 BBa_K592011]] | cjBlue |
| [[http://partsregistry.org/Part:BBa_K864401 BBa_K864401]] | aeBlue |
| [[http://partsregistry.org/Part:BBa_K592009 BBa_K592009]] | amilCP, blue chromoprotein |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414000 BBa_K414000]] | RD29A Promoter + Strong Plant Kozak (RBS) + RFP |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414006 BBa_K414006]] | 35S Promoter + (Strong Plant RBS + GFP) From K414001 |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414007 BBa_K414007]] | DREB1C promoter |
| [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K678001 BBa_K678001]] | alcA promoter |
| [[http://partsregistry.org/Part:BBa_K243004 BBa_K243004]] | Short linker |
| [[http://partsregistry.org/Part:BBa_K243005 BBa_K243005]] | Middle linker |
| [[http://partsregistry.org/Part:BBa_K243006 BBa_K243006]] | Long Linker |
| [[http://partsregistry.org/Part:BBa_K243029 BBa_K243029]] | GSAT Linker |
| [[http://partsregistry.org/Part:BBa_K243030 BBa_K243030]] | SEG Linker |
Sunday, May 5th
Picking of the plated E. coli containing biobricks and the parts received from LMU
Investigator: Florian
Aim of the experiment: The colonies from the plates were transferred into liquid LB medium, so that minipreps can be performed the next day.
Operational sequence:
- Tubes with 5 ml LB medium and the corresponding antibiotic were prepared.
- A single colony from each plate was transferred into a tube with a pipet tip.
- The biobricks were:
| Label | Name | Function |
|---|---|---|
| p19 | mCherry in pSB1C3 | |
| p20 | [[http://partsregistry.org/Part:BBa_K823039 BBa_K823039]] | GFP in pSB1C3 |
| p21 | [[http://partsregistry.org/Part:BBa_K823029 BBa_K823029]] | mKate2 in pSB1C3 |
| p22 | [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414006 BBa_K414006]] | 35S Promoter + (Strong Plant RBS + GFP) From K414001 |
| p23 | [[http://partsregistry.org/Part:BBa_K414002 BBa_K414002]] | constitutive promoter CaMV35S |
| p24 | [[http://partsregistry.org/Part:BBa_K909009 BBa_K909009]] | cDNA of UV-B sensing protein UVR8 from Arabidopsis thaliana |
| p25 | [[http://partsregistry.org/Part:BBa_K399001 BBa_K399001]] | mStrawberry |
| p26 | [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414007 BBa_K414007]] | DREB1C promoter |
| p27 | [[http://partsregistry.org/Part:BBa_K863000 BBa_K863000]] | bpul (laccase from Bacillus pumilus) with T7 promoter, RBS and HIS tag |
| p28 | [[http://partsregistry.org/Part:BBa_K337013 BBa_K337013]] | constitutive promoter SV40 |
| p29 | [[http://partsregistry.org/Part:BBa_K592011 BBa_K592011]] | cjBlue |
| p30 | [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K678001 BBa_K678001]] | alcA promoter |
| p31 | [[http://partsregistry.org/Part:BBa_K592012 BBa_K592012]] | eforRed |
| p32 | [[http://partsregistry.org/Part:BBa_K592010 BBa_K592010]] | amilGFP |
| p33 | [[http://partsregistry.org/Part:BBa_K864401 BBa_K864401]] | aeBlue |
| p34 | [[http://partsregistry.org/Part:BBa_K404319 BBa_K404319]] | mCFP |
| p35 | [[http://partsregistry.org/wiki/index.php?title=Part:BBa_K414000 BBa_K414000]] | RD29A Promoter + Strong Plant Kozak (RBS) + RFP |
| p36 | [[http://partsregistry.org/Part:BBa_K592009 BBa_K592009]] | amilCP, blue chromoprotein |
| p37 | [[http://partsregistry.org/Part:BBa_K747096 BBa_K747096]] | constitutive promoter CMV |
| p38 | [[http://partsregistry.org/Part:BBa_K147002 BBa_K147002]] | XylE dioxygenase (catechol dioxygenase) |
| p39 | [[http://partsregistry.org/Part:BBa_K243005 BBa_K243005]] | Middle linker |
| p40 | [[http://partsregistry.org/Part:BBa_K243029 BBa_K243029]] | GSAT Linker |
| p41 | [[http://partsregistry.org/Part:BBa_E0040 BBa_E0040]] | GFPmut3b |
| p42 | [[http://partsregistry.org/Part:BBa_K243006 BBa_K243006]] | Long Linker |
| p43 | [[http://partsregistry.org/Part:BBa_K243030 BBa_K243030]] | SEG Linker |
| p44 | [[http://partsregistry.org/Part:BBa_K243004 BBa_K243004]] | Short linker |
| p45 | [[http://partsregistry.org/Part:BBa_K157004 BBa_K157004]] | Fluoresceine A -binding derivative of a Lipocalin |
| p46 | [[http://partsregistry.org/Part:BBa_E2060 BBa_E2060]] | mCherry |
| p47 | [[http://partsregistry.org/Part:BBa_E2020 BBa_E2020]] | Cerulean |
| p48 | [[http://partsregistry.org/Part:BBa_E1010 BBa_E1010]] | mRFP1 |
| p49 | [[http://partsregistry.org/Part:BBa_E2050 BBa_E2050]] | mOrange |
- There were no colonies on the plate with K864401 (p33), therefore the LB medium could not be infected
- The tubes were placed into the incubator at 37 °C
Week 3
Monday, May 6th
Miniprep of the prepared E. coli containing biobricks and the parts from LMU
Investigators: Florian, Johanna, Jefferey
Aim of the experiment: A miniprep of the overnight culture of our E. coli containig our biobricks and LMU parts was performed using the Qiagen miniprep kit.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- Resulting concentrations were determined by measuring the absorption:
| Label | Name | Concentration [ng/µl] |
|---|---|---|
| p19 | mCherry in pSB1C3 | 40,2 |
| p20 | GFP in pSB1C3 | 58,5 |
| p21 | mKate2 in pSB1C3 | 131,3 |
| p22 | 35S Promoter + (Strong Plant RBS + GFP) From K414001 | 42,8 |
| p23 | constitutive promoter CaMV35S | 39,3 |
| p24 | cDNA of UV-B sensing protein UVR8 from Arabidopsis thaliana | 30,0 |
| p25 | mStrawberry | 46,6 |
| p26 | DREB1C promoter | 34,8 |
| p27 | bpul (laccase from Bacillus pumilus) with T7 promoter, RBS and HIS tag | 52,5 |
| p28 | constitutive promoter SV40 | 58,7 |
| p29 | cjBlue | 44,4 |
| p30 | alcA promoter | 26,9 |
| p31 | eforRed | 64,8 |
| p32 | amilGFP | 59,3 |
| p34 | mCFP | 78,4 |
| p35 | RD29A Promoter + Strong Plant Kozak (RBS) + RFP | 61,2 |
| p36 | amilCP, blue chromoprotein | 89,2 |
| p37 | constitutive promoter CMV | 83,6 |
| p38 | XylE dioxygenase (catechol dioxygenase) | 57,4 |
| p39 | Middle linker | 64,3 |
| p40 | GSAT Linker | 109,6 |
| p41 | GFPmut3b | 118,1 |
| p42 | Long Linker | 133,0 |
| p43 | SEG Linker | 107,5 |
| p44 | Short linker | 60,2 |
| p45 | Fluoresceine A -binding derivative of a Lipocalin | 82,7 |
| p46 | mCherry | 230,2 |
| p47 | Cerulean | 25,7 |
| p48 | mRFP1 | 299,3 |
| p49 | mOrange | 181,2 |
Sequencing of FluA, SV40, CaMV35S, xylE, LMU mKate2, LMU GFP, LMU mCherry, DREB1C, RD29A
Investigators: Andreas, Florian, Johanna, Jefferey, Rosario
Aim of the experiment: Sequencing of genes for which no prior sequencing results were available.
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode |
|---|---|---|
| p45 | FluA | FR01002355 |
| p28 | SV40 | FR01002356 |
| p23 | CaMV35S | FR01002357 |
| p38 | xylE | FR01002358 |
| p21 | LMU mKate2 | FR01002359 |
| p20 | LMU GFP | FR01002360 |
| p19 | LMU mCherry | FR01002345 |
| p46 | mCherry | FR01002346 |
| p26 | DREB1C | FR01002347 |
| p35 | RD29A | FR01002348 |
Preparation of ordered primers
Investigators: Andreas, Jefferey, Rosario
Aim of the experiment: Dissolving of primer pallets to provide a concentration of 100 pmol/µl
Procedure: We added an amount of water which gave us a final primer concentration of 100 pmol/µl.
Primers were labeled as follows:
| Label | Oligonr. |
|---|---|
| 6 | EreA_for |
| 7 | EreA_rev |
| 8 | EreA_SDM1_for |
| 9 | EreA_SDM1_rev |
| 10 | EreA_SDM2_for |
| 11 | EreA_SDM2_rev |
| 12 | EreA_SDM3_for |
| 13 | EreA_SDM3_rev |
| 14 | EreB_for |
| 15 | EreB_rev |
| 16 | EreB_SDM1_for |
| 17 | EreB_SDM1_rev |
| 18 | EreB_SDM2_for |
| 19 | EreB_SDM2_rev |
| 20 | EreB_SDM3_for |
| 21 | EreB_SDM3_rev |
Tuesday, May 7th
PCR, purification and analytical geleletrophoresis of EreA (P15) and EreB (P16)
Investigator: Jeff, Louise, Florian, Rosario, Andi, Johanna
Aim of the experiment: PCR of EreA (P15) and EreB (P16).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P15: O46 (EreA_for); P16: O31 (EreB_for)) |
| 1 µl | 10 µM Reverse Primer (P15: O47 (EreA_rev); P16: O32 (EreB_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P15; P16) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The gradient PCR program was performed after following scheme with following conditions (Tm=54 °C; ΔG=2 °C; P15 in row 11(=52 °C); P16 in row 1 (=54.0 °C)):
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| Tm=54 °C; ΔG=2 °C | 60 s | |
| 68 °C | 80 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- 9 µl of the purified PCR products were mixed with 1 µl DNA loading buffer (10x) for analytical gelelectrophoresis
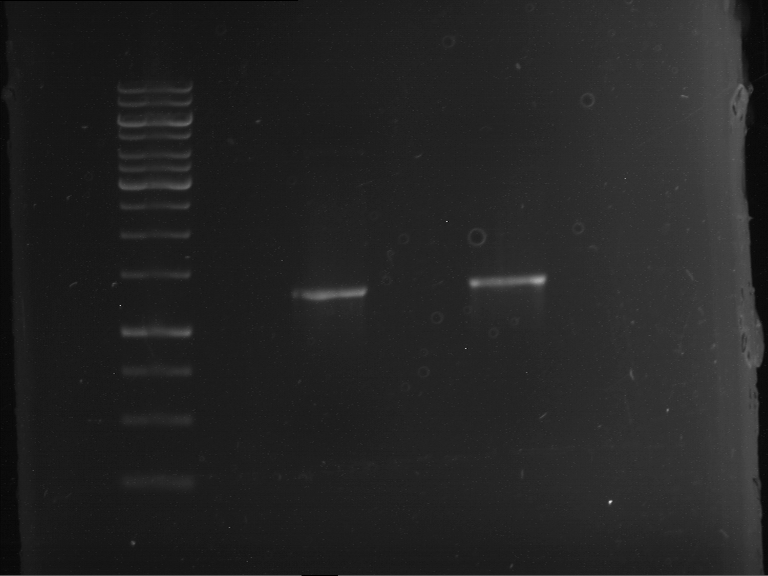
- Analytical gelelectrophoresis was performed at 90 V for 60 min in 1% agarose gel.
| 1 kbp ladder | F1 | F2 |
| Fragment-size like expected | Fragment-size like expected |
Preparative digestion and gelelectrophoresis of PhyB (P9) and PIF3-20aa linker (P50) with AgeI and PstI as well as 20aalinker-PIF3 (P51) with EcoRI and NgoMIV
Investigator: Jeff, Andi, Florian, Rosario, Louise, Johanna
Aim of the experiment: Preparative digestion and gelelectrophoresis of PhyB (P9) and PIF3-20aa linker (P50) with AgeI and PstI as well as 20aalinker-PIF3 (P51) with EcoRI and NgoMIV .
Procedure:
- Batch for preparative digestion of PhyB (P9) and PIF3-20aa linker (P50) with AgeI and PstI
| volume | reagent |
| 20 µl | Plasmid DNA P9/P50 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | PstI-HF |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of 20aalinker-PIF3 (P51) with EcoRI and NgoMIV
| volume | reagent |
| 20 µl | Plasmid DNA P51 |
| 4 µl | NEBuffer 4 |
| 1 µl | EcoRI-HF |
| 1 µl | NgoMIV |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- 4 µl of DNA loading buffer (10x) were added to the reaction batches after digestion and were loaded on an 1% low-melting agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 70 V for 90 min.
| P9 digestion = F3 | 1 kbp ladder | P50 digestion = F4 | P51 digestion = F5 |
| Fragment-size like expected; band was cut out | Fragment-size like expected; band was cut out | Fragment-size like expected; band was cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Wednesday, May 8th
Preparative digestion and gelelectrophoresis of PCR product of EreA (F1) and EreB (F2) with XbaI & AgeI
Investigator: Jeff, Andi, Florian, Rosario, Louise
Aim of the experiment: Preparative digestion and gelelectrophoresis of PCR product of EreA (F1) and EreB (F2) with XbaI & AgeI.
Procedure:
- Batch for preparative digestion of PCR product of EreA (F1) and EreB (F2) with XbaI & AgeI.
| volume | reagent |
| 25 µl | PCR product F1/F2 |
| 5 µl | NEBuffer 4 |
| 0.5 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 17.5 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of P52 for preperation of pSB1C3 vector:
| volume | reagent |
| 20 µl | Plasmid DNA P52 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- 5 µl of DNA loading buffer (10x) were added to the reaction batches for F1 & F2 and 4 µl of DNA loading buffer (10x) were added to the reaction batches for Pxx after digestion and were loaded on an 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 90 V for 60 min.
| F1 digestion = F6 | F2 digestion = F7 | 1 kbp ladder | P52 digestion = F8 |
| Length was like expected | Length was like expected | Length was like expected; the lower band was extracted |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Ligation of F8+F6 (EreA in pSB1C3), F8+F7 (EreB in pSB1C3)
Investigator: Jeff, Andi, Florian, Rosario, Louise
Aim of the experiment: Ligation of F8+F6 (EreA in pSB1C3), F8+F7 (EreB in pSB1C3). Quickchanges have still to be performed.
Procedure:
Ligation batch for F8+F6 (EreA in pSB1C3), F8+F7 (EreB in pSB1C3):
- Ligation batch for F8+F6
| volume | reagent |
| 1.35 µl | F8 (74.3 ng/µl, 2086 bp) |
| 6.76 µl | F6 (25.9 ng/µl, 1218 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 8.89 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F8+F7
| volume | reagent |
| 1.35 µl | F8 (74.3 ng/µl, 2086 bp) |
| 8.78 µl | F7 (20.6 ng/µl, 1257 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 6.87 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was als prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batches.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 100 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Midiprep of cloning vector RFC25 RFP generating device
Investigators: Andreas, Rosario
Aim of the experiment: Midiprep of our cloning vector RFC25 RFP generating device.
Procedure: Midiprep was performed after manufacturer's protocol (QIAprep Midiprep, QIAGEN)
The yield of our vector was 1854 ng.
Thursday, May 9th
Transformation of E. coli XL1 blue with F8(pSB1C3)+F6(EreA) and F8(pSB1C3)+F7(EreB)
Investigator: Florian, Jeff, Leonie, Louise, Katrin, Ingmar
Aim of the experiment: Transformation of E. coli XL1 blue with F8(pSB1C3)+F6(EreA) and F8(pSB1C3)+F7(EreB). Quickchanges has now to be performed to remove forbidden restriction sites.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of ligation products F8+F6, F8+F7, F8 negative control were added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Friday, May 10th
Quick Change mutagenesis to remove mutations found by sequencing in our gene constructs
Investigator: Andi, Jeff, Rosario
Aim of the experiment: Deletion of found mutations.
Operational sequence
1. PCR general setup
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 1 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | DNA template |
| 1 µl | 1:10 dilution of appropriate Oligo (= 5 pmol) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 12 | 95 °C | 30 s |
| 55 °C | 1 min | ||
| 68 °C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- After the PCR was finished 1 µl of the DpnI restriction enzyme (10 U/µl)was added
- Now the reaction was mixed gently and thoroughly, spinned down in a microcentrifuge for 1 minute, and immediately incubated at 37 °C for 1 hour to digest the parental supercoiled dsDNA
2. Used constructs and primers
| Quickchange number | Plasmid number | Geneconstruct | Oligo number |
| QC 1 | P27 | BPul_Laccase+ pSB1C3 | O26 & O27 |
Picking of transformed Plasmids F8 + F6
Investigator: Andi
Aim of the study: Picking from the overnight transformation of the ligated F8+F6, to see whether ligation is successful or not.
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
Investigator: Andi
PCR of TEV (P14, pRK793)
Investigator: Katrin, Jeff
Aim of the experiment: PCR, purification and analytical geleletrophoresis of TEV (P14, pRK793) to produce TEV with RFC25 pre- and suffx (still contains forbidden SpeI-site).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (O34 (TEV_fw)) |
| 1 µl | 10 µM Reverse Primer (O35 (TEV_rv)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P14) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme with following conditions:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 55 °C | 60 s | |
| 68 °C | 150 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- Resulting product was labeled as F12.
Digestion of P61 with S1 Nuclease
Investigator: Katrin
Oligohybridization of O22 (MinMCS_for) and O23 (MinMCS_rev)
Investigator: Leonie
Preparation of ordered primers
Investigators: Andreas, Rosario, Katrin, Ingmar
Aim of the experiment: Dissolving of primer pallets to provide a concentration of 100 pmol/µl
Procedure: We added an amount of water which gave us a final primer concentration of 100 pmol/µl.
Primers were labeled as follows:
| Label | Oligonr. |
|---|---|
| 24 | BPU_for |
| 25 | BPU_rev |
| 26 | BPU_SDM1_for |
| 27 | BPU_SDM1_rev |
| 28 | BPU_SDM2_for |
| 29 | BPU_SDM2_rev |
| 30 | BPU_SDM3_for |
| 31 | BPU_SDM3_rev |
| 32 | BPU_SDM4_for |
| 33 | BPU_SDM4_rev |
| 34 | TEV_for |
| 35 | TEV_rev |
| 36 | TEV_t387c_for |
| 37 | TEV_t387c_rev |
| 38 | N_Split_TEV_for |
| 39 | N_Split_TEV_rev |
| 40 | C_Split_TEV_for |
| 41 | C_Split_TEV_rev |
| 42 | PIF6_2-100_for |
| 43 | PIF6_2-100_rev |
| 44 | p104AgeI a71g fw |
| 45 | p104AgeI a71g rev |
| 46 | xylE_for |
| 47 | xylE_rev |
| 48 | xylE_c312a_for |
| 49 | xylE_c312a_rev |
| 50 | xylE_g483a_for |
| 51 | xylE_g483a_rev |
| 52 | xylE_a834g_for |
| 53 | xylE_a834g_rev |
Saturday, May 11th
Preparative digestion of PCR product of S219V pRK793 (F12) with XbaI & AgeI
Investigator: Louise
Aim of the experiment: Preparative digestion of PCR product of S219V pRK793 (F12) with XbaI & AgeI. Quickchange has still to be performed (forbidden SpeI restriction site).
Procedure:
- Batch for preparative digestion of PCR product S219V pRK793 (F12) with XbaI & AgeI.
| volume | reagent |
| 20 µl | PCR product F12 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 3 h at 37 °C
- Digested products were purified to remove nucleotides and enzymes with QIAquick PCR Purification Kit, QIAGEN.
- The digested fragment was labelled F15
Miniprep of overnight culture of transformated E.~coli XL1-Blue with F8 (pSB1C3) + F6 (EreA)
Investigator: Louise
Aim of the experiment: Miniprep of overnight culture of transformated E.~coli XL1-Blue with F8 (pSB1C3) + F6 (EreA) of 2 clones picked the day before
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
- The plasmids were labelled as P63 and P64
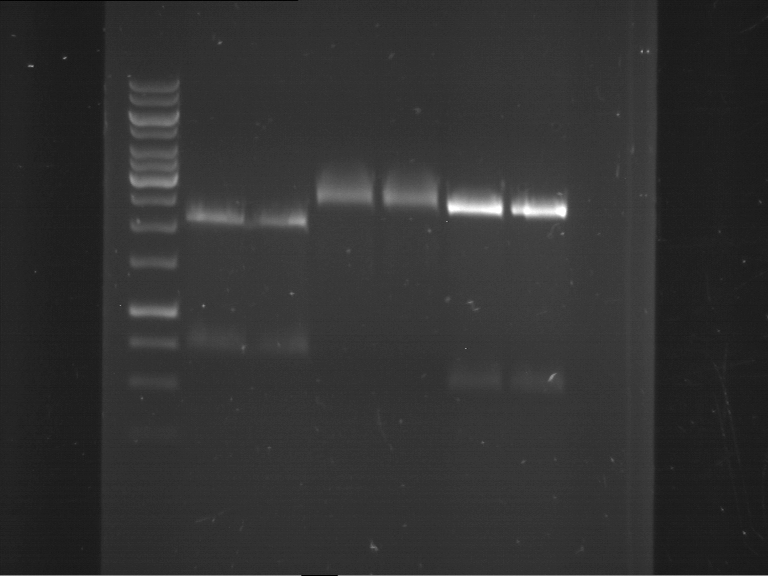
Analytical digestion and gelelectrophoresis of P63 and P64 with AgeI-HF and XbaI
Investigator: Louise
Aim of the experiment: Analytical digestion and gelelectrophoresis of P63 and P64 with AgeI-HF and XbaI
Procedure:
- Batch for analytical digestion of P63 and P64 with AgeI-HF and XbaI
| volume | reagent |
| 2.5 µl | Plasmid DNA P63/P64 |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | XbaI(10 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| Analytical digestion of P63 with AgeI-HF and XbaI | 1 kbp ladder | Analytical digestion of P64 with AgeI-HF and XbaI |
| Insertion successful | Insertion successful |
Preparative digestion of PCR product of EreB (F2) with XbaI & AgeI
Investigator: Louise,Leonie,Johanna
Aim of the experiment: Preparative digestion of PCR product EreB (F2) with XbaI & AgeI (second try since there did not grow any transformed bacteria in the first time).
Procedure:
- Batch for preparative digestion of PCR product and EreB (F2) with XbaI & AgeI.
| volume | reagent |
| 20 µl | PCR product F2 |
| 5 µl | NEBuffer 4 |
| 0.5 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 22.5 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C
- The digested fragment was labelled F14
Purification and ligation of digested PCR product EreB (F14)
Investigator: Louise, Rosario
Aim of the experiment: PCR Purification and ligation of digested PCR product EreB (F14).
Procedure: PCR Purification of EreB PCR (F14)
- To purify the digested Fragment(F14) the QIAquick PCR Purification Kit was used. The concentration of the purified product was found to be 5 ng/µl via Nano drop.
Ligation of F14 + F8(pSB1C3)
- Ligation batch for F8+F14 (positive control)
| volume | reagent |
| 1.35 µl | F8 (74.3 ng/µl, 2086 bp) |
| 15.65 µl | F14 (5 ng/µl, 1257 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
Lid temperature = 37 °C
Transformation of ligation products of F10 + F11 into E.coli Xl1-Blue
Investigator: Andi
Aim of the experiment:Transformation of the ligation products of F10 + F11 and the negative control in pTUM104 in XL1 Blue.
Operation Sequence
- melting of 2x 200 µl Ca-competent E.coli XL1-Blue cells on ice
- Preparing 4 Tubes with 100 µl of Ca-competent E.coli XL1-Blue cells
- addition of 5 µl of the ligation products
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Transformation of Quickchange Product P62 into E.coli Xl1-Blue
Investigator: Andi
Aim of the experiment:Transformation of the Quickchange Product P62 and the negative control in XL1 Blue.
Operation Sequence
- melting of 1x 200 µl Ca-competent E.coli XL1-Blue cells on ice
- Preparing 4 Tubes with 50 µl of Ca-competent E.coli XL1-Blue cells
- addition of 1 µl of the Quickchange Product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Quickchange mutagenesis of pTUM104
Investigator: Ingmar
Aim of the experiment: Sequencing results showed a forbiden AgeI restriction site in this vector. The sdm will delete this restricition site
Procedure:
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P6 template |
| 0.5 µl | 1:10 dilution of O44 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P6 template |
| 0.5 µl | 1:10 dilution of O45 (10 pmol/µL) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C. The resulting product was named fragmet 14.
Transformation into E.coli Xl1-Blue Operation Sequence
- melting of 100 µl Ca-competent E.coli XL1-Blue cells on ice
- addition of 1 µl of the PCR product
- incubation for 30 min on ice
- heat shock for 5 min at 37 °C
- transfer of cells to 1 ml LB-medium without antibiotics and incubate at 37°C and 180 rpm for 30 min
- plate 100 µl on an Amp-LB-plate
- sediment the leftover in a centrifuge (30 - 60 sec, 13 000 rpm) and resuspend the sediment in 100 µl LB-medium and plate it as well on an Amp-LB-plate
Sunday, May 12th
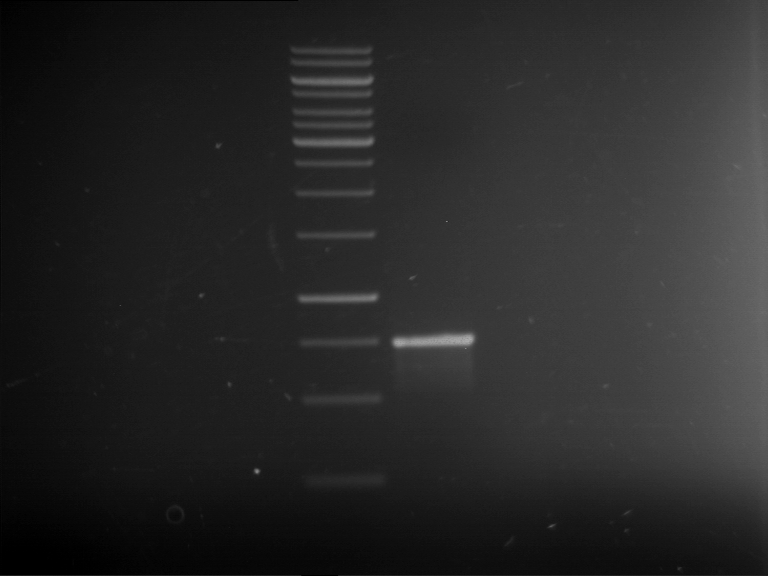
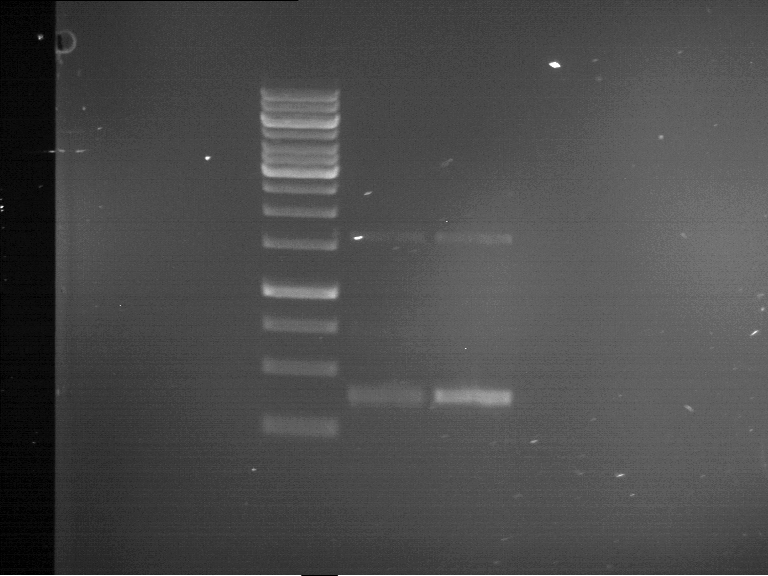
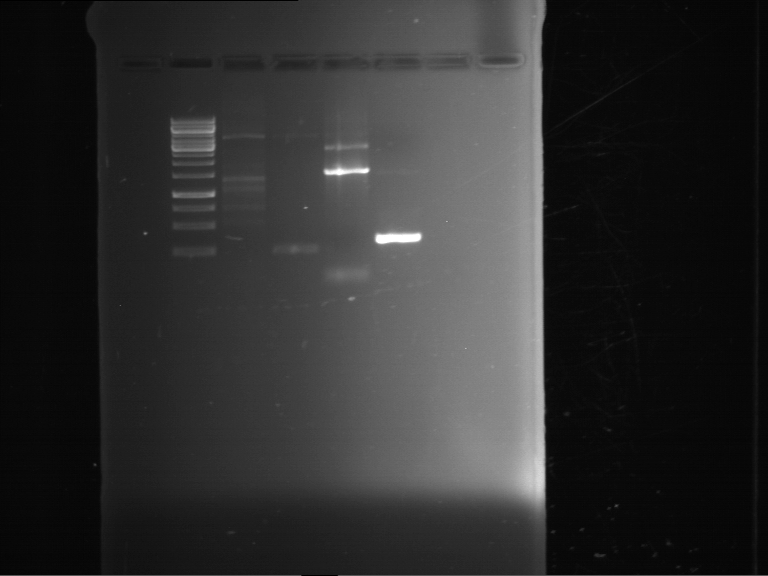
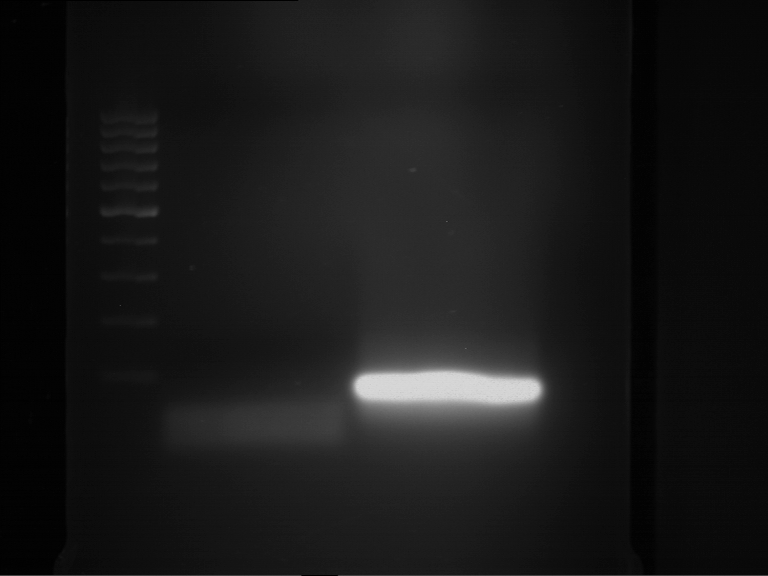
Analytical gelelectrophoresis of F15 (digested TEV PCR product)
Investigator: Jeff, Rosario
Aim of the experiment: Analytical gelelectrophoresis of F15 (digested TEV PCR product) to check whether PCR has worked.
Procedure:
- 5 µl of F15 was mixed with 0.55 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 60 min at 90 V.
- PCR product has expected length
Transformation of E. coli XL1 blue with ligation product F8+F15 (TEV in pSB1C3, RFC25)
Investigator: Jeff, Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with ligation product F8+F15 (TEV in pSB1C3, RFC25). Further quickchanges still have to be done.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of the ligation products were added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Transformation of E. coli XL1 blue with ligation products F8+F14 (EreB in pSB1C3, RFC25)
Investigator: Jeff, Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with ligation product F8+F14 (EreB in pSB1C3, RFC25). Further quickchanges still have to be done.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of the ligation products were added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Transformation of E. coli XL1 blue with P23
Investigator: Jeff, Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with P23 for insertion of miniMCS.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Picking of transformed Plasmid F10+F11 (IRES from polio virus, blunt ligated with pSB1C3 for sequencing)
Investigator: Andi
Aim of the study: Picking of transformed Plasmid F10+F11 (AlcR blunt ligated with pSB1C3 for sequencing).
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 10 colonies were picked.
Week 4
Monday, May 13th
Miniprep of overnight culture of transformated E.~coli XL1-Blue with F10+F11
Investigator: Rosario, Jeffery, Johanna, Flo, Leonie
Aim of the experiment: Miniprep of overnight culture of transformated E.~coli XL1-Blue F10+F11 of 10 clones picked the day before
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
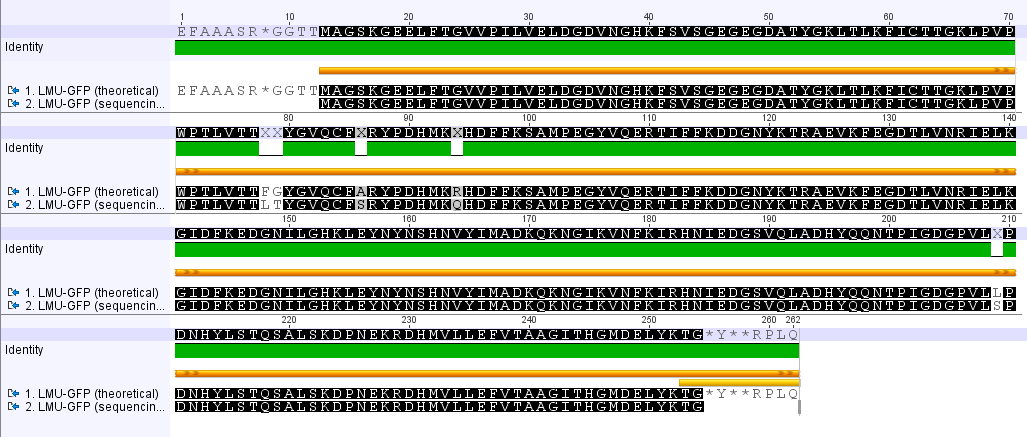
Analyzing sequencing results
Investigator: Leonie,Johanna
Aim of the experiment: Comparison of the sequencing results with the Biobrick (BB) sequences.
Procedure: Using Geneious to align the sequencing results with the BB sequences.
LMU GFP in RFC 25(p20)
- The provided sequence in the parts registry is totally wrong.
- The comparison of the amino acid seqeunces reveals several point mutations.
FluA in RFC 25 (p45)
- Did not yield sequencing results (does the primer bind ?)
- No presence of RCF 25 standard restriction enzymes
mCherry in RFC 25 (p46)
- Did not yield sequencing results (does the primer bind ?)
- No presence of RCF 25 standard restriction enzymes
CaMV35S in RFC 25 (p23)
- high identity
- high match with HQ699544 (Blast)
LMU mKate2in RFC 25 (p21)
- no continous ORF
- Did not yield sequencing results
DREB1C promoter in RFC 25 (p26)
- Present of fluorecent protein gene
xylE in RCF 25 (p38)
- Did not yield sequencing results
SV40 in RCF 25 (p28)
- sequencing result agrees with the laccases
- the sequencing result can't be the one of SV40
Quickchange mutagenesis SDMI of p27
Investigator: Rosario, Jeff, Flo, Andi
Aim of the experiment: Sequencing results showed a forbidden restriction site in this vector. The sdm will delete this restricition site. The resulting Plasmid was labeled 65.
Procedure:
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0.5 µl | Plasmid P27 template |
| 0.5 µl | 1:10 dilution of O26 (10 pmol/µL) |
| 20. 5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0.5 µl | Plasmid P27 template |
| 0.5 µl | 1:10 dilution of O27 (10 pmol/µL) |
| 20.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 4 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied, but the amount of cycles was increased to 20
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Quickchange mutagenesis SDMI of p38
Investigator: Jeff, Flo, Andi, Rosario
Aim of the experiment: Sequencing results showed a forbidden restriction site in this vector. The sdm will delete this restricition site. The resulting Plasmid was labeled 67.
Procedure:
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0,5 µl | Plasmid P38 template |
| 0.5 µl | 1:10 dilution of O48 (10 pmol/µL) |
| 20.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0.5 µl | Plasmid P38 template |
| 0.5 µl | 1:10 dilution of O49 (10 pmol/µL) |
| 20.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 4 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied, but the amount of cycles have been increased to 20
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Quickchange mutagenesis SDMI of P63
Investigator: Jeff, Rosario, Flo, Andi
Aim of the experiment: Sequencing results showed a forbidden restriction site in this vector. The sdm will delete this restricition site. The resulting Plasmid was labeled P66.
Procedure:
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0.25 µl | Plasmid P63 template; 1:2 dilution |
| 0.5 µl | 1:10 dilution of O8 (10 pmol/µL) |
| 20.75 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 0.25 µl | Plasmid P63 template; 1:2 dilution |
| 0.5 µl | 1:10 dilution of O9 (10 pmol/µL) |
| 20.75 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 10 | 95°C | 30 sec |
| 55°C | 1 min | ||
| 67°C | 6 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied, the amount of cycles was raised to 20
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the PCR batch and incubate for 1 h at 37 °C.
Transformation of E. coli XL1 blue with P65
Investigator: Jeff, Rosario, Johanna, Andi, Flo, Leonie
Aim of the experiment: Transformation of E. coli XL1 blue with P65 .
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Transformation of E. coli XL1 blue with P66
Investigator: Jeff, Rosario, Flo, Johanna, Andi, Leonie
Aim of the experiment: Transformation of E. coli XL1 blue with P66.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Transformation of E. coli XL1 blue with P67
Investigator: Jeff, Rosario, Johanna, Andi, Leonie, Flo
Aim of the experiment: Transformation of E. coli XL1 blue with P67.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Tuesday, May 14th
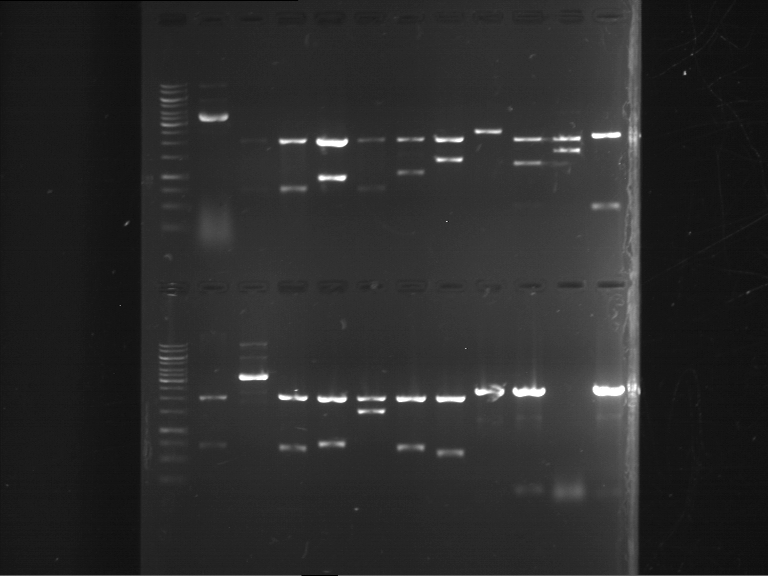
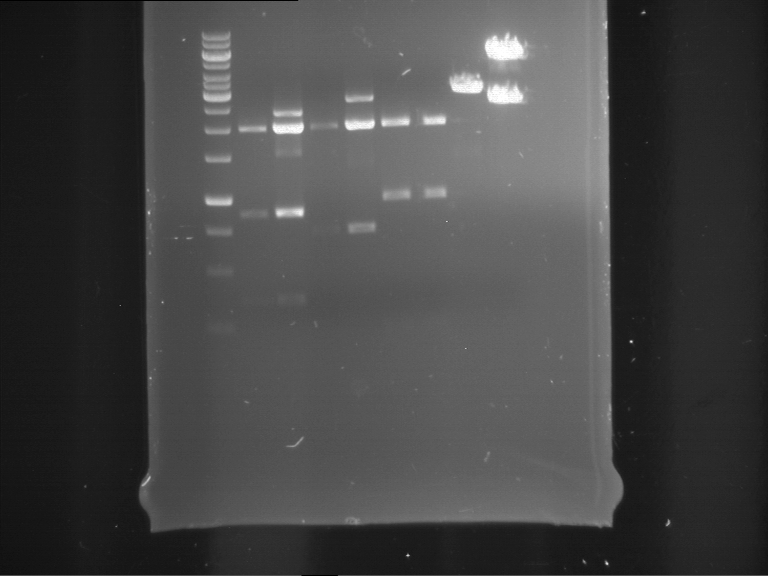
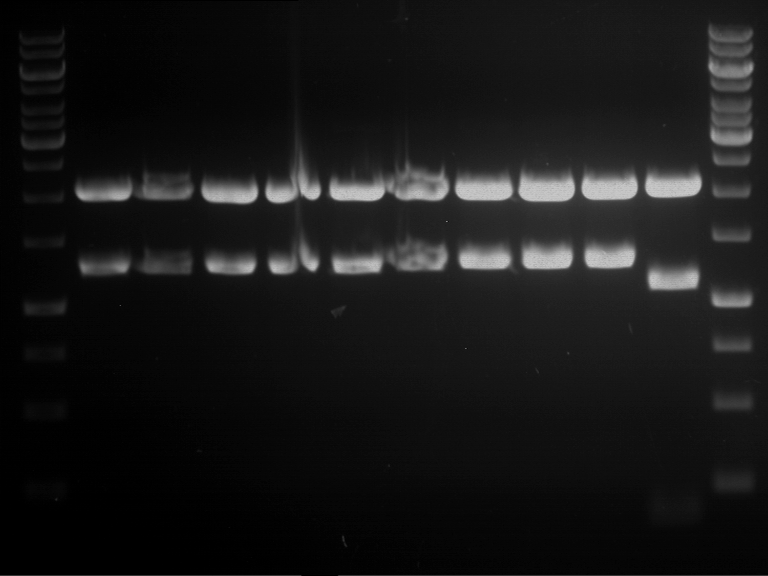
Analytical digestion of P15 and P19 to P52
Investigator: Leonie, Florian, Johanna
Aim of the experiment: Analytical digestion and gelelectrophoresis of P15 and P19 to P52 to check the sequencing results.
Procedure:
- Batch for analytical digestion of P15 and P19 to P52
| volume | reagent |
| 5 µl | Plasmid DNA |
| 5 µl | Mastermix |
| =10 µl | TOTAL |
Batch of Mastermix
| volume | reagent |
| 40 µl | NEBuffer 4 (10x) |
| 4 µl | BSA (100x) |
| 0.2 µl | NotI(10 U/µl) |
| 148 µl | ddH2O |
| =200 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
- Discussion:
- Gel 1, Row 1:
| lane | part | plasmid | band 1 | band 2 | result |
| 2 | EreA | pDEST14 | 3,25 kb | no cut, no restriction sites in pDEST14 | |
| 3 | mCherry(LMU) | pSB1C3 | 2,0 kb | 750 bp | probably right |
| 4 | GFP(LMU)(845 bp) | pSB1C3 | 2,0 kb | 750 bp | maybe switched with mKate2(LMU) |
| 5 | mKate2(LMU)(702 bp) | pSB1C3 | 2,0 kb | 900 bp | maybe switched with GFP(LMU) |
| 6 | 35S + Strong Plant + GFP(1775 bp) | pSB1C3 | 2,0 kb | 750 bp | ? |
| 7 | UVR8(1332 bp) | pSB1C3 | 2,0 kb | 1,1 kb | ? |
| 8 | mStrawberry(708 bp) | pSB1C3 | 2,0 kb | 1,4 kb | ? |
| 9 | DREB1C promoter(463 bp) | pSB1C3 | 2,5 kb | probably only one cut -> one NotI site mutated | |
| 10 | Laccase(1587 bp) | pSB1C3 | 2,0 kb | 1,3 kb | ? |
| 11 | SV40(419 bp) | pSB1C3 | 2,0 kb | 1,6 kb | this could be the laccase |
| 12 | cjBlue(696 bp) | pSB1C3 | 2,0 kb | 0,5 kb | ? |
- Gel 1, Row 2:
| lane | part | plasmid | band 1 | band 2 | result |
| 2 | alcA (843 bp) | pSB1C3 | 2,0 kb | 0,7 kb | ? |
| 3 | eforRed (681 bp) | pSB1C3 | plasmid | plasmid bands (coiled, supercoiled, etc.) | |
| 4 | amilGFP (699 bp) | pSB1C3 | 2,0 kb | 0,7 kb | right part |
| 5 | mCFP (714 bp) | pSB1C3 | 2,0 kb | 0,7 kb | right part |
| 6 | RD29A promoter (1535 bp) | pSB1C3 | 2,0 kb | 1,5 kb | right part |
| 7 | amilCP (669 bp) | pSB1C3 | 2,0 kb | 0,7 kb | right part |
| 8 | CMV promoter (580 bp) | pSB1C3 | 2,0 kb | 0,6 kb | right part |
| 9 | middle linker (24 bp) | pMA-BBFR | 2,4 kb | short insert went through the gel | |
| 10 | GSAT (108 bp) | pMA-BBFR | 2,4 kb | 0,1 kb | right part |
| 11 | GFP(720 bp) | pSB1A2 | ? | ||
| 12 | long linker (36 bp) | pMA-BBFR | 2,4 kb | short insert went through the gel |
- Gel 2:
| lane | part | plasmid | band 1 | band 2 | result |
| 2 | SEG linker (108 bp) | pMA-BBFR | 2,4 kb | short insert went through the gel | |
| 3 | short linker (12 bp) | pMA-BBFR | 2,4 kb | short insert went through the gel | |
| 4 | FluA (522 bp) | pMA-BBFR | 2,4 kb | 0,5 kb | right part |
| 5 | mCherry (769 bp) | pSB2K3 | 4,4 kb | 0,75 kb | right part |
| 6 | Cerulean (778 bp) | pSB2K3 | ? | ||
| 7 | mRFP1 (706 bp) | pSB2K3 | 4,4 kb | 0,7 kb | right part |
| 8 | mOrange (769 bp) | pSB2K3 | 4,4 kb | 0,75 kb | right part |
| 9 | PIF3-20aa (366 bp) | pSB1C3 | 2,0 kb | 0,35 kb | right part |
| 10 | 20aa-PIF3 (366 bp) | pSB1C3 | 2,0 kb | 0,35 kb | two plasmids and two insert can be seen |
| 11 | LexA (ca. 6 kb) | pSB1C3 | > 8 kb | only one cut, huge plasmid |
Picking of Plasmid P23 (CaMV35S) and Ligation Product F8 + F15 (TEV)
Investigator: Jeff, Florian
Aim of the study: Picking of Plasmid P23 (CaMV35S) and Ligation Product F8 + F15 (TEV) for Miniprep.
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 1 colony of P23 were picked.
- 7 colonies of F8 + F15 were picked.
Sequencing of P63 & P64
Investigators: Louise
Aim of the experiment: Sequencing of genes for which no prior sequencing results were available.
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| p63 | EreA | FR01002349 | FR01002350 |
| p64 | EreA | FR01002351 | FR01002352 |
Wednesday, May 15th
Preparative digestion of psB1C3-RFP-generator (P60) and another psB1C3 (P8) as well as P64 (EreA in defect psB1C3 (F8)) with XbaI & AgeI
Investigator: Louise,Leonie,Johanna
Aim of the experiment:Preparative digestion of psB1C3-RFP-generator (P60) and another psB1C3 (P8) as well as P64 (EreA in defect psB1C3 (F8)) with XbaI & AgeI to recover EreA and prepare two new vectors for cloning.
Procedure:
- Batch for preparative digestion of PCR product and EreB (F2) with XbaI & AgeI.
| volume | reagent |
| 20 µl | Plasmid (P8,P60,P64) |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C
- The digested fragments were labelled F17(P8), F18(P60), F19(P64)
PCR, purification and analytical geleletrophoresis of and EreB (P16)
Investigator: Jeff, Louise
Aim of the experiment: PCR of EreB (P16).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (O31 (EreB_for)) |
| 1 µl | 10 µM Reverse Primer (O32 (EreB_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P15; P16) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program TMKS was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 52 °C | 60 s | |
| 68 °C | 80 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- 4 µl of the purified PCR products were mixed with 0.4 µl DNA loading buffer (10x) for analytical gelelectrophoresis
- Analytical gelelectrophoresis was performed at 90 V for 30 min in 1% agarose gel.
| 1 kbp ladder | F16 |
| Fragment-size like expected |
Purification and ligation of digested products F17, F18, F19
Investigator: Jeff, Louise
Aim of the experiment: Purification and ligation of digested products F17, F18, F19.
Procedure:
- To purify the digested Fragments the QIAquick PCR Purification Kit was used. The concentration of the purified product was found to be XX ng/µl via Nano drop.
Ligation of F19 + FXX(pSB1C3)
- Ligation batch for F19+FXX (positive control)
| volume | reagent |
| 1.35 µl | F8 (74.3 ng/µl, 2086 bp) |
| X µl | F14 (5 ng/µl, 1257 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
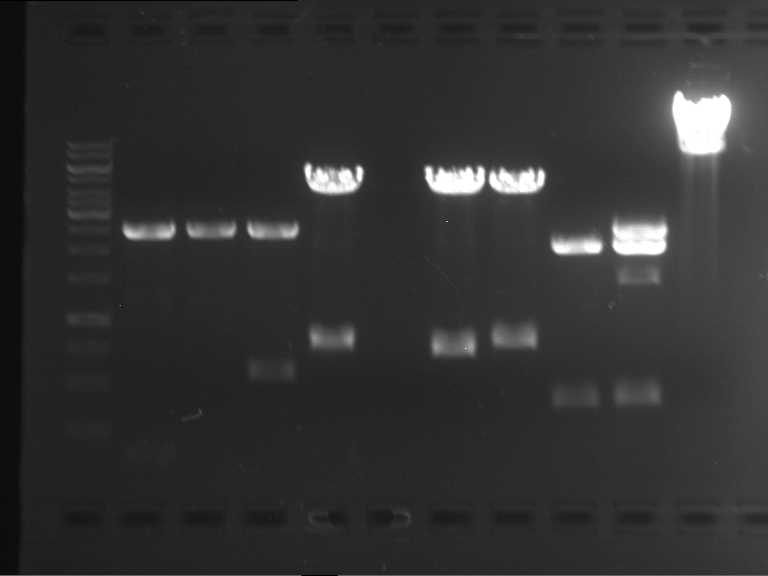
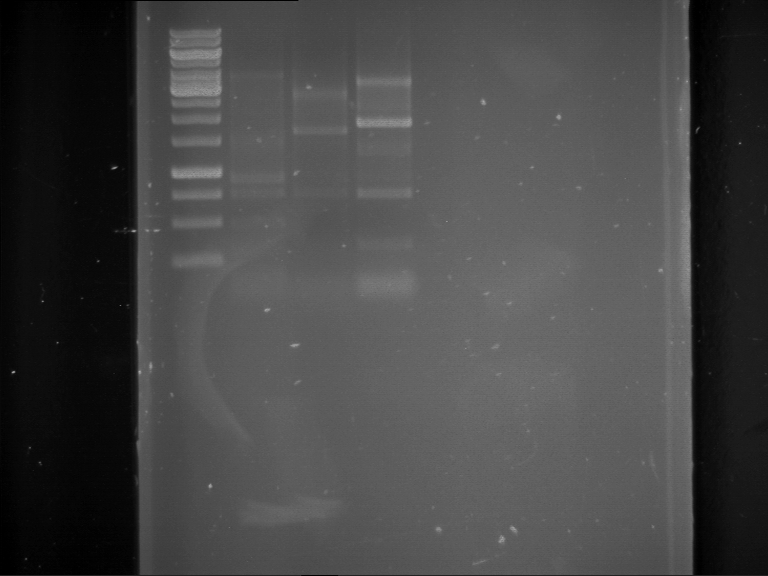
Analytical digestion and analytical gelelectrophoresis of P90-P97 (TEV protease, XbaI+AgeI) and P97 (CaMV p35S, XbaI+PstI)
Investigator: Jeff
Aim of the experiment: Analytical digestion and analytical gelelectrophoresis of P90-P97 (TEV protease, XbaI+AgeI) and P97 (CaMV p35S, XbaI+PstI).
Procedure:
- Mastermix for analytical digestion of P90-P96 with XbaI+AgeI-HF:
| volume | reagent |
| 16 µl | NEBuffer 4 (10x) |
| 1.6 µl | BSA (100x) |
| 2 µl | XbaI (20 U/µl) |
| 2 µl | AgeI-HF (20 U/µl) |
| 118.4 µl | ddH2O |
| =140 µl | TOTAL |
- Batch for digestion of P90-P96 with XbaI+AgeI-HF:
| volume | reagent |
| 2.5 µl | Plasmid DNA |
| 17.5 µl | Mastermix |
| =20 µl | TOTAL |
- Batch for digestion of P90-P96 with XbaI+AgeI-HF:
| volume | reagent |
| 2.5 µl | Plasmid DNA |
| 2 µl | NEBuffer 4 (10x) |
| 0.2 µl | BSA (100x) |
| 0.25 µl | XbaI (20 U/µl) |
| 0.25 µl | AgeI-HF (20 U/µl) |
| 14.8 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | Digestion of P90 | Digestion of P91 | Digestion of P92 | Digestion of P93 | Digestion of P94 | Digestion of P95 | Digestion of P96 | Digestion of P97 |
| Insert has expected length | Insert has expected length | Insert has expected length | Insert has expected length | Corrupt | Insert has expected length | Insert has expected length | Corrupt |
- The inserts for TEV semm to be right in the most cases, but the backbone has mutated and it should be cloned into a new pSB1C3 with new same restriction enzymes (XbaI+AgeI)!
- The forward sequencing of CaMV p35S is right but the analytical digestion is wrong. Maybe further parts are downstream of the promoter?! Reverse sequencing should be performed for further analysis.
Thursday, May 16th
Preparative digestion of pSH21 (P61), psB1A2(P41) and P96 (TEV S19V)
Investigator: Jeff, Katrin
Aim of the experiment: Preparative digestion of pSH21 (P61)and psB1A2(P41) with EcoRI and P96 (TEV S19V) with XbaI & AgeI for subsequent ligation.
Procedure:
- Batch for preparative digestion of pSH21 with EcoRI
| volume | reagent |
| 20 µl | Plasmid DNA P61 |
| 4 µl | Buffer 4(10x) |
| 1 µl | EcoRI (20 U/µl) |
| 15 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of psB1A2 with EcoRI
| volume | reagent |
| 20 µl | Plasmid DNA P41 |
| 4 µl | Buffer 4(10x) |
| 1 µl | EcoRI (20 U/µl) |
| 15 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of P96 (TEV S219V) with XbaI & AgeI.
| volume | reagent |
| 20 µl | Plasmid DNA P96 |
| 4 µl | Buffer 4(10x) |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- The resulting fragments were named: F23 (P41) and F22 (P61)
- 4 µl of DNA loading buffer (10x) were added to the reaction batch containing pSH21 (P61) after digestion and were loaded on an 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 70 V for 90 min.
| 1 kbp ladder | P61 digestion = F23 |
| Fragment-size like expected; band was cut out |
- The band had the expected length. They were cut out and purified using the QIAquick Gel Extraction Kit, QIAGEN
Preparative digestion of ereB (P64) with XbaI & AgeI.
Investigator: Jeff, Louise, Katrin
Aim of the experiment:Preparative digestion of EreB (P64) with XbaI & AgeI.
Procedure:
- Batch for preparative digestion of PCR product and EreB (F2) with XbaI & AgeI.
| volume | reagent |
| 20 µl | Plasmid |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 13.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C
- The digested fragments were labelled
Dephosphorylation of F23
Investigator: Jeff, Katrin
Aim of the experiment: Dephosphorylation of cut vector psB1A2 to prevent re-ligation
Procedure:
- the digestion product was purified with QIAquick PCR Purification Kit, Qiagen in order to change the buffer
- Batch for the dephosphorylation of F23
| volume | reagent |
| 30 µl | Plasmid DNA F23 (2 µg) |
| 4 µl | 10X Fast AP buffer |
| 2 µl | FastAP Thermosensitive Alkaline
Phosphatase |
| 4 µl | ddH2O |
| =40 µl | TOTAL |
- the reaction batch was spun briefly and incubated at 37 °C for 10 minutes
- the reaction was stopped by heating to 75 °C for 5 minutes
Ligation of pSH21 (F22) with psB1A2(F23), TEV (P96), EreA (P64), pSB1A2(P41)
Investigator: Jeff, Katrin
Aim of the experiment: ligation of pSH21 (F22) with pSB1A2(F23), TEV (F20) with pSB1C3 (F17), EreA (F19)with pSB1C3 (F17) and EreB (F21) with pSB1C3 (F17)
Procedure:
- Ligation batch for EreB (F21) with pSB1C3 (F17)
| volume | reagent |
| 1.6 µl | F17 (61,4 ng/µl) |
| 15,4 µl | F21 (8,3 ng/µl) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| =20 µl | TOTAL |
- Ligation batch for TEV (F20) with pSB1C3 (F17)
| volume | reagent |
| 1.6 µl | F17 (61,4 ng/µl) |
| 4,1 µl | F20 (24,7 ng/µl) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 11,3 µl | ddH20 |
| =20 µl | TOTAL |
- Ligation batch for pSH21 (F22) with pSB1A2(F23)
| volume | reagent |
| 0,85 µl | F23 (118,1 ng/µl) |
| 2,3 µl | F22 (119,0 ng/µl) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 13,85 µl | ddH20 |
| =20 µl | TOTAL |
- Ligation batch for EreA (F19) with pSB1C3 (F17)
| volume | reagent |
| 1.6 µl | F17 (61,4 ng/µl) |
| 6,4 µl | F19 (8,3 ng/µl) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 9 µl | ddH20 |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- the ligation was performed at 16 °C overnight
Friday, May 17th
Transformation of E. coli XL1 blue with F17+F19, F17+F20, F17+F21, F22+F23, F17 (negative control), F23 (negative control)
Investigator: Jeff, Katrin
Aim of the experiment: Transformation of ligation products F17+F21 (pSB1C3, EreB), F17+F20 (pSB1C3, TEV), F17+F19 (pSB1C3, EreA), F23+F22 (pSB1A2, pSH21) and negative controls
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on chloramphenicol plates (F17+F19, F17+F20, F17+F21, F17 negative) or ampicillin plates (F23+F22, F23 negative control)
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was discarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates (F17+F19, F17+F20, F17+F21) or ampicillin plates (F23+F22)
Quick Change mutagenesis to remove mutations found by sequencing in our gene constructs
Investigator: Jeff, Leonie
Aim of the experiment: removal of forbidden restiction sites from laccase
Operational sequence
PCR general setup
| volume | reagent |
| 5 µl | 10x Pfu Ultra II buffer |
| 1 µl | DNA template (p28) |
| 0,5 µl each | 1:10 dilution of appropriate Oligos (1: O26 + O27, 2: O28 + O29, 3: O30 + O31, 4: O32 + O33 ) |
| 42 µl | ddH2O |
| 1 µl | dNTP mix |
| 1 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 sec |
| 2 | 12 | 95 °C | 30 s |
| 55 °C | 1 min | ||
| 68 °C | 4 min | ||
| 3 | 1 | 4 °C | hold |
Transformation of E. coli XL1 blue with the 4 Quickchangeproducts using 1: O26 + O27, 2: O28 + O29, 3: O30 + O31, 4: O32 + O33
Investigator: Jeff, Andi, Johanna, Leonie
Aim of the experiment: Transformation of E. coli XL1 blue with the 4 Quickchangeproducts using 1: O26 + O27, 2: O28 + O29, 3: O30 + O31, 4: O32 + O33
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on chloramphenicol plates 1: O26 + O27, 2: O28 + O29, 3: O30 + O31, 4: O32 + O33
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was discarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on chloramphenicol plates (1: O26 + O27, 2: O28 + O29, 3: O30 + O31, 4: O32 + O33)
Saturday, May 18th
Picking of transformed Plasmid E. coli XL1 blue with F17+F19, F17+F20, F17+F21, F22+F23
Investigator: Andi, Jeff
Aim of the study: Picking of transformed Plasmid F17+F19, F17+F20, F17+F21, F22+F23 .
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR/AmpR LB-medium)
- 2 colonies were picked for every Transformation product.
Sunday, May 19th
Miniprep of overnight culture of transformated E. coli XL1 blue with F17+F19, F17+F20, F17+F21, F22+F23
Investigator: Jeff, Andi
Aim of the experiment: Miniprep of overnight culture of transformated E. coli XL1 blue with F17+F19, F17+F20, F17+F21, F22+F23 of 2 clones picked the day before
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
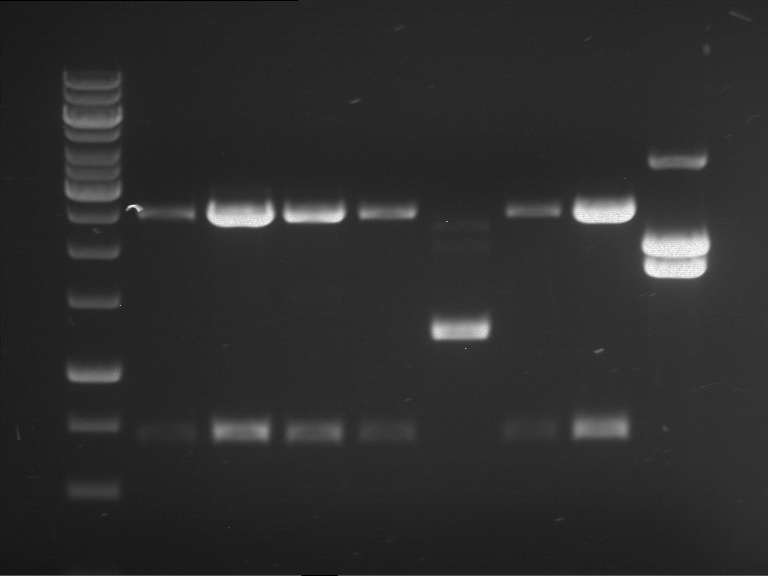
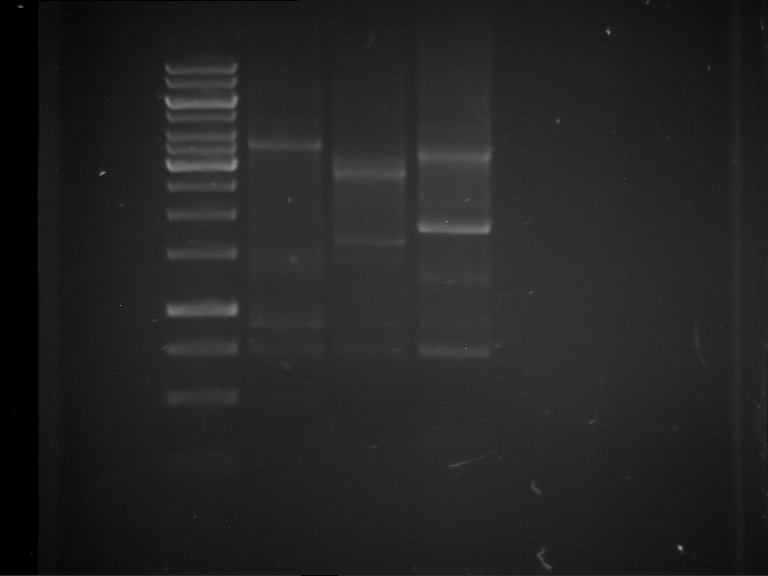
Analytical digestion and analytical gelelectrophoresis of F17+F19, F17+F20, F17+F21 and F22+F23
Investigator: Jeff, Andi
Aim of the experiment: Analytical digestion and analytical gelelectrophoresis of ligation products F17+F19, F17+F20, F17+F21, F22+F23.
Procedure:
- Mastermix for analytical digestion of F17+F19, F17+F20, F17+F21 with XbaI+PstI-HF:
| volume | reagent |
| 14 µl | NEBuffer 4 (10x) |
| 1.4 µl | BSA (100x) |
| 1.75 µl | XbaI (20 U/µl) |
| 1.75 µl | AgeI-HF (20 U/µl) |
| 103.6 µl | ddH2O |
| =122.5 µl | TOTAL |
- Batch for digestion of F22+F23 with EcoRI-HF:
| volume | reagent |
| 2.5 µl | Plasmid DNA |
| 2 µl | NEBuffer 4 (10x) |
| 15.25 µl | ddH2O |
| 0.25 µl | EcoRI-HF (20 U/µl) |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed two times for each ligation product at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | Digestion of F17+F19 | Digestion of F17+F19 | Digestion of F17+F20 | Digestion of F17+F20 | Digestion of F17+F21 | Digestion of F17+F21 | Digestion of F22+F23 | Digestion of F22+F23 |
| Insert has expected length | Insert has expected length, the first line represents the rest of the uncut Plasmid | Insert has expected length | Insert has expected length, the first line represents the rest of the uncut Plasmid | Insert has expected length | Insert has expected length | Insert has expected length, insert and cut Plasmid overlay each other, because of the same length | Corrupt |
Quick Change mutagenesis to remove forbidden restriction sites of P28 (Laccase), P100 (TEV S219V), P102 (EreB)
Investigator: Jeff, Andi
Aim of the experiment: Removal of forbidden restiction sites from P28 (Laccase), P100 (TEV S219V), P102 (EreB).
Operational sequence
PCR general setup
Procedure:
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid template (P28/P100/P102) |
| 1 µl | 1:10 dilution of O26 (for P28, 10 µM)/O16 (for P102, 10 µM)/O36 (for P100, 10 µM) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P38 template |
| 1 µl | 1:10 dilution of O27 (for P28, 10 µM)/O17 (for P102, 10 µM)/O37 (for P100, 10 µM) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 10 | 95 °C | 30 s |
| 55 °C | 1 min | ||
| 68 °C | 4 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied, but the amount of cycles have been increased to 20.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Analytical gelelectrophoresis of QuickChange products of P100, P102 and P28 to check whether the QuickChange PCR and DpnI digestion were successful
Investigator:
Aim of the experiment:
Procedure:
Transformation of the Quickchange products with P28, P100 and P102 into E. coli XL1 blue
Investigator: Jeff, Andi
Aim of the experiment: Transformation of E. coli XL1 blue with P28, P102 and P100.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Week 5
Tuesday, May 21st
Quick Change mutagenesis to remove forbidden restriction sites - SDMI - of P28 (Laccase), P100 (TEV S219V), P102 (EreB)
Investigator: Jeff, Andi, Johanna
Aim of the experiment: Removal of forbidden restiction sites from P28 (Laccase), P100 (TEV S219V), P102 (EreB).
Operational sequence
PCR general setup
Procedure:
PCR
Reaction batch 1
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid template (P28/P100/P102) |
| 1 µl | 1:10 dilution of O26 (for P28, 10 µM)/O16 (for P102, 10 µM)/O36 (for P100, 10 µM) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch 2
| volume | reagent |
| 2.5 µl | 10x Pfu Ultra II buffer |
| 4 µl | Plasmid P38 template |
| 1 µl | 1:10 dilution of O27 (for P28, 10 µM)/O17 (for P102, 10 µM)/O37 (for P100, 10 µM) |
| 16.5 µl | ddH2O |
| 0.5 µl | dNTP mix |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 10 | 95 °C | 30 s |
| 55 °C | 1 min | ||
| 68 °C | 4 min |
- Having completed the PCR cycling parameters listed above both PCR reaction batches were mixed together and the cycling parameters listed above were one time more applied, but the amount of cycles have been increased to 20.
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Analytical gelelectrophoresis of QuickChange products of P100, P102 and P28 to check whether the QuickChange PCR and DpnI digestion were successful
Investigator:
Aim of the experiment:
Procedure:
Transformation of the Quickchange products with P28, P100 and P102 into E. coli XL1 blue
Investigator: Jeff, Andi, Johanna
Aim of the experiment: Transformation of E. coli XL1 blue with P28, P102 and P100.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Sequencing of P105, P28, P100, P102 & P98
Investigators: Jeff, Andi,Johanna
Aim of the experiment: Sequencing of genes for which no prior sequencing results were available.
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| p105 | IRES | FR01002288 | FR01002269 |
| p102 | EreB | FR01002270 | FR01002275 |
| p100 | TEV S219V | FR01002278 | FR01002279 |
| p98 | EreA | FR01002276 | FR01002277 |
| p28 | Laccase | FR01002280 |
Preparation of Knop-Medium for Moss
Investigator: Jeff, Andi, Johanna, Rosario
Aim of the experiment: Preparation of Knop-Medium for Moss
Procedure:
- All four prepared stocks have been: 25g/L KH2PO4; 25g/L KCl; 25g/L MgSO4 x 7 H2O; 100g/L Ca(NO3)2
- Volume of prepared Medium: 1L
- 10 ml of each stock were converted into a 1,8L Erlenmeyer flask and filled up to 1L with destilled water (ELGA); 12,5mg FeSO4 x 7 H2O were added; the pH was set to 5.8 with KOH
- The prepared Volume of 1L was shared equally to 500ml Erlenmeyer flasks, so each Erlenmeyer flask contained 200 ml of the Knop-Medium
- All flasks have been autoclaved
Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P100 (TEV, QC with O36/O37) and P102 (EreB, QC with O16/O17)
Investigator: Andi, Jeff, Johanna, Rosario
Aim of the study: Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P100 (TEV, QC with O36/O37) and P102 (EreB, QC with O16/O17) from Sunday, May 19th.
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 3 colonies for P100QC (TEV, QC with O36/O37) and 5 colonies from P102QC (EreB, QC with O16/O17) were picked from the plates.
Wednesday, May 22nd
Miniprep of overnight culture of transformated E. coli XL1 blue with QuickChange products of P100 (TEV, QC with O36/O37) and P102 (EreB, QC with O16/O17)
Investigator: Jeff
Aim of the experiment: Miniprep of overnight culture of transformated E. coli XL1 blue with QuickChange products of P100 (TEV, QC with O36/O37) and P102 (EreB, QC with O16/O17).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P28 (Laccase, QC with O26/O27)
Investigator: Jeff
Aim of the study: Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P28 (Laccase, QC with O26/O27).
Operational sequence:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 8 colonies for P28QC (Laccase, QC with O26/O27) were picked from the plates.
Analytical digestion and analytical gelelectrophoresis of P106, P107, P108, P109, P110, P111, P112, P113
Investigator: Jeff, Rosario, Leonie
Aim of the experiment: Analytical digestion and analytical gelelectrophoresis of Quickchanges to check whether the transformation of E. coli Xl1 Blue with QuickChanges products of P100 (TEV, QC with O36/O37: P111, P112, P113) and P102 (EreB, QC with O16/O17: P106, P107, P108, P109, P110) were successful
Procedure:
Mastermix for analytical digestion of P106, P107, P108, P109, P110, P111, P112, P113 with AgeI-HF and XbaI:
| volume | reagent |
| 20 µl | NEBuffer 4 (10x) |
| 2 µl | BSA (100x) |
| 2.5 µl | AgeI-HF (20 U/µl) |
| 2.5 µl | XbaI |
| 148 µl | ddH2O |
| = 175 µl | TOTAL |
- 17,5 µl Mastermix with 2,5 µl Plasmid
Mastermix for analytical digestion of P111, P112, P113 with XbaI and SpeI and P106, P107, P108, P109, P110 with XbaI and PstI-HF:
| volume | reagent |
| 20 µl | NEBuffer 4 (10x) |
| 2 µl | BSA (100x) |
| 2.5 µl | XbaI |
| 148 µl | ddH2O |
| = 172,5 µl | TOTAL |
- 17,25 µl Mastermix with 2,5 µl Plasmid and 0,25 µl SpeI or PstI respectively
Mastermix for analytical digestion of with P106, P107, P108, P109, P110 EcoRI and SpeI:
| volume | reagent |
| 12 µl | NEBuffer 4 (10x) |
| 1,2 µl | BSA (100x) |
| 1,5 µl | EcoRI-HF |
| 1,5 µl | SpeI-HF |
| 88.8 µl | ddH2O |
| = 105 µl | TOTAL |
- 17,5 µl Mastermix with 2,5 µl Plasmid
Mastermix for analytical digestion of with P106, P107, P108, P109, P110 XbaI and SpeI:
| volume | reagent |
| 12 µl | NEBuffer 4 (10x) |
| 1,2 µl | BSA (100x) |
| 1,5 µl | XbaI |
| 1,5 µl | SpeI-HF |
| 88.8 µl | ddH2O |
| = 105 µl | TOTAL |
- 17,5 µl Mastermix with 2,5 µl Plasmid
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder | P111 XbaI + AgeI | P112 XbaI + AgeI | P113 XbaI + AgeI | P111 XbaI + SpeI | P112 XbaI + SpeI | P113 XbaI + SpeI |
| Ctrl | Ctrl | Ctrl | QC succesful | QC failed | QC succesful |
| 1 kbp ladder DNA ladder | P106 XbaI + AgeI | P107 XbaI + AgeI | P108 XbaI + AgeI | P109 XbaI + AgeI | P110 XbaI + AgeI | P106 XbaI + PstI | P107 XbaI + PstI | P108 XbaI + PstI | P109 XbaI + PstI | P110 XbaI + PstI |
| Ctrl | Ctrl | Ctrl | Ctrl | Ctrl | PstI inside as should be; useless experiment | PstI inside as should be; useless experiment | PstI inside as should be; useless experiment | PstI inside as should be; useless experiment | PstI inside as should be; useless experiment |
| 1 kbp ladder DNA ladder | P106 XbaI + SpeI | P107 XbaI + SpeI | P108 XbaI + SpeI | P109 XbaI + SpeI | P110 XbaI + SpeI | P106 EcoRI + SpeI | P107 EcoRI + SpeI | P108 EcoRI + SpeI | P109 EcoRI + SpeI | P110 EcoRI + SpeI |
| Ctrl | Ctrl | Ctrl | Ctrl | Ctrl | QC succesful | QC succesful | QC succesful | QC succesful | QC failed |
Thursday, May 23rd
Sequencing of P111, P113 and P109 (?)
Investigator: Jeff
Aim of the experiment: Sequencing of P111, P113 and P109 to check whether the QuickChange brought further unwanted mutations.
Procedure:
PCR of P111 (TEV S219V, SpeI-less) to generate Split-TEV
Investigator: Jeff
Aim of the experiment: PCR of P111 (TEV S219V, SpeI-less) to generate Split-TEV. To generate N-TEV, the primer O38 and O39 were used. To generate C-TEV, the primer O40 and O41 were used.
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (O38 (N_Split_TEV_fw) for generating N-TEV; O40 (C_Split_TEV_fw) for generating C-TEV) |
| 1 µl | 10 µM Reverse Primer (O39 (N_Split_TEV_rv) for generating N-TEV; O41 (C_Split_TEV_rv) for generating C-TEV) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P111) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme with following conditions:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 51 °C | 60 s | |
| 68 °C | 30 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
- Resulting product was labeled as F24 (N-TEV) and F25 (C-TEV).
Analytical gelelectrophoresis of F24 and F25 (N-TEV and C-TEV)
Investigator: Jeff, Rosario
Aim of the experiment: Analytical gelelectrophoresis of F24 and F25 (N-TEV and C-TEV) to check whether PCR has worked.
Procedure:
- 4 µl of F24 and F25 were mixed with seperately with 0.44 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 60 min at 90 V.
- PCR products has expected length, but there are unspecific products. So we have to be cautious and have to check if we ligate the right products.
Preparative digestion of PCR products (N-TEV and C-TEV) of P111 (TEV S219V, SpeI-less) with XbaI & AgeI
Investigator: Jeff, Rosario
Aim of the experiment: Preparative digestion of PCR products (N-TEV and C-TEV) of P111 (TEV S219V, SpeI-less) with XbaI & AgeI to clone it into pSB1C3.
Procedure:
- Batch for preparative digestion of PCR products (N-TEV and C-TEV) of P111 (TEV S219V, SpeI-less) with XbaI & AgeI.
| volume | reagent |
| 25 µl | PCR product F24/F25 |
| 5 µl | NEBuffer 4 |
| 0.5 µl | BSA (100x) |
| 1 µl | AgeI-HF |
| 1 µl | XbaI |
| 17.5 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 150 min at 37 °C.
- Digested products were purified with QIAquick PCR Purification Kit, QIAGEN.
- The digested fragments were labelled F26 (digestion of F24 with XbaI&AgeI) and F27 (digestion of F25 with XbaI&AgeI)
Ligation of F17+F26 (N-TEV in pSB1C3) and F17+F27 (C-TEV in pSB1C3)
Investigator: Jeff, Rosario
Aim of the experiment: Ligation of F17+F26 (N-TEV in pSB1C3) and F17+F27 (C-TEV in pSB1C3).
Procedure:
- Ligation batch for F17+F26:
| volume | reagent |
| 1.63 µl | F17 (61.4 ng/µl, 2086 bp) |
| 3.20 µl | F26 (15.9 ng/µl, 354 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.17 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F17+F27 (positive control)
| volume | reagent |
| 1.63 µl | F17 (61.4 ng/µl, 2086 bp) |
| 2.41 µl | F27 (21.1 ng/µl, 354 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.96 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
- Lid temperature = 37 °C
Miniprep of overnight culture of transformated E. coli XL1 blue with the first Quickchange (SDMI) Product of P28
Investigator: Jeff, Andi, Rosario, Johanna, Leonie
Aim of the experiment: Miniprep of overnight culture of transformated E. coli XL1 blue with the first Quickchange Product of P28. 8 clones were picked the day before.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and analytical gelelectrophoresis of P28, P117, P118, P119, P120, P121, P122, P123
Investigator: Rosario, Andi, Leonie, Johanna, Jeff
Aim of the experiment: Analytical digestion and analytical gelelectrophoresis of ligation products P28, P117, P118, P119, P120, P121, P122, P123.
Procedure:
- Mastermix for analytical digestion of P28, P117, P118, P119, P120, P121, P122, P123 AgeI-HF:
| volume | reagent |
| 20 µl | NEBuffer 4 (10x) |
| 2 µl | BSA (100x) |
| 2.5 µl | AgeI-HF (20 U/µl) |
| 150.5 µl | ddH2O |
| =175 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed two times for each ligation product at 90 V for 60 min.
Results:
| 1 kbp ladder DNA ladder | Digestion of P28 | Digestion of P116 | Digestion of P117 | Digestion of P118 | Digestion of P119 | Digestion of P120 | Digestion of P121 | Digestion of P122 | Digestion of P122 |
| to be announced | to be announced | to be announced | to be announced | to be announced | to be announced | to be announced | to be announced | to be announced |
Quick Change mutagenesis to remove forbidden restriction sites - SDMII - of P28 (Laccase), P106 (EreB)
Investigator: Jeff, Andi, Johanna, Rosario, Leonie
Aim of the experiment: Removal of forbidden restiction sites from P28 (Laccase), P106 (EreB).
Procedure: Quickchange - PCR
Reaction batch - P106
| volume | reagent | Additional information |
| 5 µl | 10x Pfu Ultra II buffer | |
| 2 µl | Plasmid template (P106 - 1:10 dilution) | need to get to 50 ng/µl |
| 1.25 µl | 1:10 dilution of O18 (for P106, 10 µM) | |
| 1.25 µl | 1:10 dilution of O19 (for P106, 10 µM) | |
| 38.5 µl | ddH2O | |
| 1 µl | dNTP mix | |
| 1 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P106
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 18 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Reaction batch - P116 (Laccase)
| volume | reagent | Additional information |
| 5 µl | 10x Pfu Ultra II buffer | |
| 2 µl | Plasmid template - 1:5 dilution | need to get to 50 ng/µl |
| 1.25 µl | 1:10 dilution of O28 for P116 | |
| 1.25 µl | 1:10 dilution of O29 for P116 | |
| 38.5 µl | ddH2O | |
| 1 µl | dNTP mix | |
| 1 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P116
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 18 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Transformation of the Quickchange products with P28, P100 and P102 into E. coli XL1 blue
Investigator: Jeff, Andi, Johanna, Rosario
Aim of the experiment: Transformation of E. coli XL1 blue with Quickchangeproduct SDM II of P106 and P116.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Friday, May 24th
Transformation of E. coli XL1 blue with ligation products F17+F26 (N-TEV S219V in pSB1C3) and F17+F27 (C-TEV S219V in pSB1C3)
Investigator: Jeff, Florian
Aim of the experiment: Transformation of E. coli XL1 blue with ligation products F17+F26 (N-TEV S219V in pSB1C3) and F17+F27 (C-TEV S219V in pSB1C3).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions was plated again on a new chlorampenicol plates.
Quick Change mutagenesis to remove forbidden restriction sites - SDMII - of P116 (Laccase), P109 (EreB)
Investigator: Volker, Louise, Katrin, Leonie, Flo
Aim of the experiment: Removal of forbidden restiction sites from P116 (Laccase), P109 (EreB).
Procedure: Quickchange - PCR
Reaction batch - P109
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P109 - 1:10 dilution) | need to get to 50 ng/µl |
| 1 µl | 1:10 dilution of O18 (for P109, 10 µM) | |
| 1 µl | 1:10 dilution of O19 (for P109, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P109
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 21 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min 50 s |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Reaction batch - P116 (Laccase)
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template - 1:5 dilution | need to get to 50 ng/µl |
| 1 µl | 1:10 dilution of O28 for P116 | |
| 1 µl | 1:10 dilution of O29 for P116 | |
| 18.0 µl | ddH2O | |
| 1 µl | dNTP mix 50 mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P116
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 21 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min 50 s |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Transformation of the Quickchange products into E. coli XL1 blue
Investigator: Volker
Aim of the experiment: Transformation of E. coli XL1 blue with Quickchangeproduct SDM II of P109 and P116.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Saturday, May 25st
Analytical gelelectrophoresis of QuickChange products of P116 and P109 to check whether the QuickChange PCR and DpnI digestion were successful
Investigator: Volker, Louise, Katrin, Leonie, Flo
Aim of the experiment: Analytical gelelectrophoresis of QuickChange products of P116 and P109 to check whether the QuickChange PCR and DpnI digestion were successful
Procedure:
- 5 µl of the PCR Quickchange products were either mixed with with 0.5 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 60 min at 90 V.
Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19)
Investigator: Volker
Aim of the study: Picking of transformed Plasmid E. coli XL1 blue with QuickChange products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19).
Procedure:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 5 colonies for P116QC (Laccase, QC with O28/O29) and 6 colonies for P109QC (EreB, QC with O18/O19) were picked from the plates.
Picking of E. coli XL1 blue with ligation products F17+F26 (N-TEV S219V in pSB1C3) and F17+F27 (C-TEV S219V in pSB1C3)
Investigator: Volker
Aim of the study: Picking of E. coli XL1 blue with ligation products F17+F26 (N-TEV S219V in pSB1C3) and F17+F27 (C-TEV S219V in pSB1C3).
Procedure:
- Picking and overnight culture after standard laboratory's protocol. (CamR LB-medium)
- 5 colonies were picked from plates for E. colis XL1 blue transformed with ligation products F17+F26 (N-TEV S219V in pSB1C3) and F17+F27 (C-TEV S219V in pSB1C3).
Sunday, May 26th
Miniprep of overnight cultures of transformated E. coli XL1 blue with QuickChange products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19)
Investigator: Katrin, Louise
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with QuickChange products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis to check whether the transformation of E. coli Xl1 Blue with QuickChanges products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19) were successful
Investigator: Katrin, Louise
Aim of the experiment:Analytical digestion and gelelectrophoresis to check whether the transformation of E. coli Xl1 Blue with QuickChanges products of P116 (Laccase, QC with O28/O29) and P109 (EreB, QC with O18/O19)
Procedure:
- analytical digestion of Quickchange products of P109 (EreB, QC with O18/O19)
| volume | reagent |
| 2 µl | NEB CutSmart Buffer (10x) |
| 0,25 µl | PstI-HF (20 U/µl) |
| 2,5 µl | DNA from Miniprep |
| 15,25 µl | ddH2O |
| =20 µl | TOTAL |
- in order to check if the QC was successful, P109 (before QC) was also digested
- analytical digestion of Quickchange products of P116 (Laccase, QC with O28/O29)
| volume | reagent |
| 2 µl | NEB CutSmart Buffer (10x) |
| 0,25 µl | EcoRI-HF |
| 0,25 µl | NgoMIV |
| 2,5 µl | DNA from Miniprep |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- in order to check if the QC was successful, P116 (before QC) was also digested
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder# | P116 (Laccase before QCII) digested with EcoRI-HF, NgoMIV | Laccase after QCII (P128) digested with EcoRI-HF, NgoMIV | Laccase after QCII (P129) digested with EcoRI-HF, NgoMIV | Laccase after QCII (P130) digested with EcoRI-HF, NgoMIV | Laccase after QCII (P131) digested with EcoRI-HF, NgoMIV | Laccase after QCII (P124) digested with EcoRI-HF, NgoMIV | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment |
| as expected | as expected | as expected | as expected | as expected | as expected |
Laccase P124 was chosen for sequencing
| 1 kbp ladder DNA ladder | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | EreB after QCII (P125) digested with PstI | EreB after QCII (P132) digested with PstI | EreB after QCII (P133) digested with PstI | EreB after QCII (P134) digested with PstI | EreB after QCII (P135) digested with PstI | EreB after QCII (P136) digested with PstI | EreB before QCII (P109) digested with PstI |
| as expected | as expected | as expected | as expected | as expected | as expected | as expected |
EreB P125 was chosen for sequencing
Miniprep of ligation products of N-TEV (F17+F26) and C-TEV (F17+F25) with pSB1C3
Investigator: Katrin, Louise
Aim of the experiment: Miniprep of ligation products of N-TEV (F17+F26) and C-TEV (F17+F25) with pSB1C3
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of ligation products F17 + F26, F17 + F27
Investigator: Katrin, Louise
Aim of the experiment: Analytical digestion and gelelectrophoresis to check whether the ligation of N-TEV (F17+F26) and C-TEV (F17+F25) in pSB1C3 were successful
Procedure:
- analytical digestion of ligation products of N-TEV (F17+F26) and C-TEV (F17+F25)in pSB1C3
Mastermix:
| volume | reagent |
| 22 µl | NEB Buffer 4 (10x) |
| 2,2 µl | BSA (100X) |
| 2,75 µl | XbaI |
| 2,75 µl | AgeI-HF |
| 162,8 µl | ddH2O |
| =192,5 µl | TOTAL |
- 2,5 µl of miniprep products were added to 17,5 µl Mastermix
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder# | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | N-TEV ligated into pSB1C3 (F17 + F26) (P126) digested with XbaI, AgeI-HF | N-TEV ligated into pSB1C3 (F17 + F26) (P137) digested with XbaI, AgeI-HF | N-TEV ligated into pSB1C3 (F17 + F26) (P138) digested with XbaI, AgeI-HF | N-TEV ligated into pSB1C3 (F17 + F26) (P139) digested with XbaI, AgeI-HF |
| as expected | as expected | as expected | as expected |
N-TEV ligated into pSB1C3 (F17 + F26) (P126) was chosen for sequencing
| 1 kbp ladder DNA ladder | C-TEV ligated into pSB1C3 (F17 + F27) (P127) digested with XbaI, AgeI-HF | C-TEV ligated into pSB1C3 (F17 + F27) (P140) digested with XbaI, AgeI-HF | C-TEV ligated into pSB1C3 (F17 + F27) (P141) digested with XbaI, AgeI-HF | C-TEV ligated into pSB1C3 (F17 + F27) (P142) digested with XbaI, AgeI-HF | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment | belongs to other experiment |
| as expected | as expected | as expected | as expected |
C-TEV ligated into pSB1C3 (F17 + F27) (P127) was chosen for sequencing
Sequencing of P124-P127
Investigators: Louise, Katrin
Aim of the experiment: Sequencing of QCII products (EreB, Laccase) and ligation product of split-TEV
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| p124 | Miniprep of transformation of QC product of P116 (Laccase, QC with O28/O29)(QC - SDMII - of Bpul Laccase to remove forbidden NgoMIV) | FR01002337 (VR) | FR01002338 (VF2) |
| p125 | Miniprep of transformation of QC product of P109 (EreB, QC with O18/O19) (QC - SDMII - of EreB to remove forbidden PstI) | FR01002339 (VR) | FR01002340 (VF2) |
| p126 | Miniprep of ligation product F17 + F26 (N-TEV) | FR01002341 (VR) | FR01002342 (VF2) |
| p127 | Miniprep of ligation product F17 + F27 (C-TEV) | FR01002343 (VR) | FR01002344 (VF2) |
Week 6
Monday, May 27th
Oligohybridization of MiniMCS (O56,O57) and Spytag (O54,O55)
Investigator: Leonie,Johanna
Aim of the experiment: Oligohybridization of MiniMCS (MiniMCSII_fw (O56) and MiniMCSII_rv (O57) and of Spytag (SpyTag_fw (O54) and SpyTag_rv (O55))
Operational sequence
- 25 µL of 100 pmol/µl of MiniMCS fw and 25 µL of 100 pmol/µl MiniMCS rev in one Eppi. 25 µL of 100 pmol/µl of Spytag fw and 25 µL of 100 pmol/µl Spytag rev in a second Eppi
- Heating up to 95 °C for 5 min
- Cooling at RT in a styropor box overnight
PCR of npt-Kasette (P115), PIF3 (P51), pActin (P114) and t35S (P114)
Investigator: Leonie, Johanna
Aim of the experiment: PCR of npt-Kasette (P115), PIF3 (P51), pActin (P114) and t35S (P114).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction GC Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P114:O62 (pActin_for); P114:O60 (t35S_for); P115:O58 (npt-Kasette_for); P51:O66 (PIF3_for)) |
| 1 µl | 10 µM Reverse Primer (P114:O63 (pActin_rev); P114:O61 (t35S_rev); P115:O59 (npt-Kasette_rev); P51:O67 (PIF3_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P114; P115; P51) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 49 °C | 60 s | |
| 68 °C | 100 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Results:
- PCR did not work properly because primer concentration was wrong.
Preparative digestion and gelelectrophoresis of P45 (FluA-Psb1C3) with XbaI & AgeI, P39 with AgeI & PstI, P20 with NgoMIV & PstI
Investigator: Andi
Aim of the experiment:Preparative digestion of P45 (FluA-Psb1C3) with XbaI & AgeI, P39 with AgeI & PstI, P20 with NgoMIV & PstI to clone fragment of P45 into F17 (pSB1C3) and to fusion Middle linker of P39 with GFP of P20.
Procedure:
Batch for preparative digestion of P45 (FluA-Psb1C3) with XbaI & AgeIwith XbaI & AgeI.
| volume | reagent |
| 20 µl | P45 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 2 µl | AgeI-HF |
| 2 µl | XbaI |
| 11.6 µl | ddH2O |
| =40 µl | TOTAL |
Batch for preparative digestion of P39 (Middle-Linker) with PstI and AgeI.
| volume | reagent |
| 20 µl | P39 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 2 µl | AgeI-HF |
| 2 µl | PstI |
| 11.6 µl | ddH2O |
| =40 µl | TOTAL |
Batch for preparative digestion of P20 (GFP-Psb1C3) with NgoMIV and PstI.
| volume | reagent |
| 20 µl | P45 |
| 4 µl | NEBuffer 4 |
| 0.4 µl | BSA (100x) |
| 2 µl | NgoMIV |
| 2 µl | PstI |
| 11.6 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 120 min at 37 °C.
- Preparative gelelectrophoresis was performed at 90 V for 1.5 h.
| 1 kbp ladder | P45 digested with XbaI&AgeI (=F28) | P45 digested with XbaI&AgeI (=F28) | P39 digested with AgeI&PstI (=F29) | P39 digested with AgeI&PstI (=F29) | P20 digested with NgoMIV&PstI (=F30) | P20 digested with NgoMIV&PstI (=F30) |
| lower band was cut out | lower band was cut out | band was cut out | band was cut out | lower band was cut out | lower band was cut out |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
Quick Change mutagenesis to remove forbidden restriction sites - SDMIII - of P124(Laccase), P125 (EreB)
Investigator: Andi
Aim of the experiment: Removal of forbidden restiction sites from P124 (Laccase), P125 (EreB).
Procedure: Quickchange - PCR
Reaction batch - P124
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P124 - 1:6 dilution) | need to get to 50 ng/µl |
| 1 µl | 1:10 dilution of O30 (for P125, 10 µM) | |
| 1 µl | 1:10 dilution of O31 (for P125, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
Reaction batch - P125
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P125 - 1:6 dilution) | need to get to 50 ng/µl |
| 1 µl | 1:10 dilution of O20 (for P125, 10 µM) | |
| 1 µl | 1:10 dilution of O21 (for P125, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters for both batches
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 21 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min 50 s |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Preparation of ordered primers
Investigators: Andi
Aim of the experiment: Dissolving of primer pallets to provide a concentration of 100 pmol/µl
Procedure: We added an amount of water which gave us a final primer concentration of 100 pmol/µl.
Primers were labeled as follows:
| Label | Oligonr. |
|---|---|
| O54 | Spytag_fw |
| O55 | Spytag_rev |
| O56 | MiniMCSII_fw |
| O57 | MiniMCSII_rev |
| O58 | Npt_for |
| O59 | Npt_rev |
| O60 | t35S_for |
| O61 | t35S_rev |
| O62 | pACT_for |
| O63 | pACT_rev |
| O64 | NucA_for |
| O65 | NucA_rev |
| O66 | Pif3_for |
| O67 | Pif3_rev |
Transformation of P45, P51, P100 and P102 into E. coli XL1 blue
Investigator: Andi, Johanna
Aim of the experiment: Transformation of E. coli XL1 blue with P45, P51, P102 and P100.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Picking of transformed Plasmid E. coli XL1 blue with P114, P115
Investigator: Andi
Aim of the study: Picking of transformed Plasmid E. coli XL1 blue with P114, P115.
Procedure:
- Picking and overnight culture after standard laboratory's protocol. (AmpR LB-medium)
- 5 colonies of P114 and P115 were picked from the plates.
Ligation of F29+F30 (GFP in pSB1C3 after middle linker, ( Gly-Gly-Ser-Gly)x2) and F17+F27 (FluA in pSB1C3)
Investigator: Jeff
Aim of the experiment: Ligation of F29+F30 (GFP in pSB1C3 after middle linker, ( Gly-Gly-Ser-Gly)x2) and F17+F27 (FluA in pSB1C3).
Procedure:
- Ligation batch for F29+F30:
| volume | reagent |
| 4.22 µl | F29 (23.7 ng/µl, 2110 bp) |
| 10.48 µl | F30 (9.5 ng/µl, ~700 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 2.3 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F17+F28:
| volume | reagent |
| 1.63 µl | F17 (61.4 ng/µl, 2086 bp) |
| 7.8 µl | F28 (9.6 ng/µl, 522 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 7.57 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
- Lid temperature = 37 °C
Tuesday, May 28th
Analytical gelelectrophoresis of PCR products of P114 (O62&O63), P114 (O60&O61), P115 (O58&O59), P51 (O66&O67)
Investigator: Jeff, Rosario, Leonie
Aim of the experiment: Analytical gelelectrophoresis of PCR products of P114 (O62&O63), P114 (O60&O61), P115 (O58&O59), P51 (O66&O67).
Procedure:
- 4 µl of PCR products of P114 (O62&O63), P114 (O60&O61), P115 (O58&O59), P51 (O66&O67) were mixed with seperately with 0.44 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 40 min at 90 V.
| 1 kbp ladder | PCR products of P114 (O62&O63), P114 (O60&O61), P115 (O58&O59), P51 (O66&O67) | PCR products of P114 (O60&O61) | PCR products of P115 (O58&O59) | PCR products of P51 (O66&O67) |
| PCR did work, but strong primer dimer signal | PCR did work, but strong primer dimer signal | PCR did not work | PCR did work, but strong primer dimer signal |
- Wrong buffer (GC buffer, instead of standard reaction buffer, was used and 10x too much primers were also used)
Transformation of Quickchangeproduct SDMIII of P124 (Laccase) and P125 (EreB) + ligation product of F29+F30 + ligation product of F17+F19 into E. coli XL1 blue
Investigator: Andi, Johanna
Aim of the experiment: Transformation of Quickchangeproduct SDMIII of P124 (Laccase) and P125 (EreB) + ligation product of F29+F30 + ligation product of F17+F19 + ligation product of F17+NK + ligation product of F29+NK into E. coli XL1 blue.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 2 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Sequencing of P133 and P134
Investigators: Louise, Johanna
Aim of the experiment: Sequencing of two more QCII products (EreB) Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| p133 | Miniprep of transformation of QC product of P109 (EreB, QC with O18/O19) (QC - SDMII - of EreB to remove forbidden PstI) | FR01002287 (fw) | FR01002296 (rev) |
| p134 | Miniprep of transformation of QC product of P109 (EreB, QC with O18/O19) (QC - SDMII - of EreB to remove forbidden PstI) | FR01002295 (fw) | FR01002294 (rev) |
Miniprep of overnight cultures of transformated E. coli XL1 blue of P114 and P115
Investigator: Andi
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue of P114 and P115.
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Picking of of E. coli XL1 blue transformed with P45, P51, P100, P102
Investigator: Jeff, Leonie, Rosario
Aim of the experiment: Picking of of E. coli XL1 blue transformed with P45, P51, P100, P102.
Procedure:
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x) (P51, P100, P102)/4 µL Ampicillin (1000x) (P45).
- 1 colony for each plasmid were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight.
PCR of npt-Kasette (P144), PIF3 (P51), pActin (P143) and t35S (P143)
Investigator: Rosario, Leonie, Jeff
Aim of the experiment: PCR of npt-Kasette (P144), PIF3 (P51), pActin (P143) and t35S (P143).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P143:O62 (pActin_for); P143:O60 (t35S_for); P144:O58 (npt-Kasette_for); P51:O66 (PIF3_for)) |
| 1 µl | 10 µM Reverse Primer (P143:O63 (pActin_rev); P143:O61 (t35S_rev); P144:O59 (npt-Kasette_rev); P51:O67 (PIF3_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P114; P115; P51) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 47 °C | 60 s | |
| 68 °C | 120 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Analytical gelelectrophoresis of F33, F34, F35, F36
Investigator: Jeff, Rosario, Leonie
Aim of the experiment: Analytical gelelectrophoresis of F33, F34, F35, F36.
Procedure:
- 4 µl of F33, F34, F35 and F36 were mixed with seperately with 0.44 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 40 min at 90 V.
| 1 kbp ladder | F33 | F33 | F33 | F33 |
| PCR did not work, maybe high GC content? | PCR products has expected length, some byproducts | PCR products has expected length, some byproducts | PCR products has expected length |
Wednesday, May 29th
Transformation of E. coli XL1 blue with SDMIII product of P124, ligation product of F29+F30 and ligation product of F17+F18
Investigator: Andi
Aim of the experiment: Transformation of E. coli XL1 blue with SDMIII product of P124, ligation product of F29+F30 and ligation product of F17+F18.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 5 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Miniprep of overnight cultures of transformated E. coli XL1 blue of P100, P102, P51, P45
Investigator: Rosario
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue of P100, P102, P51, P45 .
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Picking of of E. coli XL1 blue transformed with ligation product of F17+F28
Investigator: Andi
Aim of the experiment: Picking of of E. coli XL1 blue transformed with ligation product of F17+F28.
Procedure:
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x) (
- 3 colonies were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight.
Quick Change mutagenesis to remove forbidden restriction sites - SDMIII - of P133 (EreB)
Investigator: Louise
Aim of the experiment: Removal of forbidden restiction site from P133 (EreB).
Procedure: Quickchange - PCR
Reaction batch
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P133 - 1:10 dilution) | need to get to 50 ng/µl |
| 1 µl | 1:10 dilution of O20 (for P133, 10 µM) | |
| 1 µl | 1:10 dilution of O21 (for P133, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P133
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 19 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 3 min 30 s |
- Digestion of the parental DNA with DpnI: Addition of 1 µl DpnI to the reaction batch and incubate for 1 h at 37 °C.
Transformation of the Quickchange product into E. coli XL1 blue
Investigator: Louise
Aim of the experiment: Transformation of E. coli XL1 blue with Quickchangeproduct SDM III of P133.
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 100 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
Preparative digestion of t35s(F34) with XbaI & PstI, npt(F25) with EcoRI & SpeI, PIF3(F36) with XbaI & AgeI-HF and psB1C3 with XbaI & PstI as well as with EcoRI & SpeI
Investigator: Louise
Aim of the experiment: Preparative digestion of t35s(F34) with XbaI & PstI, npt(F25) with EcoRI & SpeI, PIF3(F36) with XbaI & AgeI-HF and psB1C3(P10) with XbaI & PstI as well as with EcoRI & SpeI.
Procedure:
- Batch for preparative digestion of t35s(F34) with XbaI and PstI
| volume | reagent |
| 25 µl | Plasmid DNA P61 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of npt(F35) with EcoRI and SpeI
| volume | reagent |
| 25 µl | Plasmid DNA P61 |
| 5 µl | CutSmart Buffer |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | SpeI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of PIF3(F36) with XbaI and AgeI-HF
| volume | reagent |
| 25 µl | Plasmid DNA P61 |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | AgeI-HF (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Batch for preparative digestion of psB1C3(P10) with XbaI & PstI.
| volume | reagent |
| 20 µl | Plasmid DNA P96 |
| 4 µl | CutSmart Buffer |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Batch for preparative digestion of psB1C3(P10) with EcoRI and SpeI.
| volume | reagent |
| 20 µl | Plasmid DNA P96 |
| 4 µl | CutSmart Buffer |
| 1 µl | EcoRI (20 U/µl) |
| 1 µl | SpeI (20 U/µl) |
| 14 µl | ddH2O |
| =40 µl | TOTAL |
- Incubation for 2.5 h at 37 °C.
- The resulting fragments were named: ?????
- 5 µl of DNA loading buffer (10x) were added to the 50 µl reaction batches and 4 µl of DNA loading buffer (10x) were added to the 40 µl reaction batches after digestion and were loaded on a 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 90 V for 90 min.
Transformation of E. coli XL1 blue with pSB1C3 containing a NLS from SV40 (BBa_K801030)
Investigator: Florian, Jeff
Aim of the experiment: Transformation of E. coli XL1 blue with pSB1C3 containing a NLS from SV40 (BBa_K801030).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA was added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellet was resuspended in 100 µl of LB-medium and this concentrated cell suspension was plated again on a new chlorampenicol plate.
PCR of pActin (P143)
Investigator: Florian, Jeff
Aim of the experiment: PCR of pActin (P143).
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer (P143:O62 (pActin_for)) |
| 1 µl | 10 µM Reverse Primer (P143:O63 (pActin_rev)) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA (P143) |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 51 °C | 60 s | |
| 68 °C | 80 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Results:
- After analytical gelelectrophoresis there was no product visible. PCR did't work!
Ligation of F17+F40 (PIF3 in pSB1C3(RFC25)), F42+F38 (t35S in pSB1C3(RFC10)) and F41+F39(npt-casette in pSB1C3(RFC10))
Investigator: Jeff
Aim of the experiment: Ligation of F17+F40 (PIF3 in pSB1C3(RFC25)), F42+F38 (t35S in pSB1C3(RFC10)) and F41+F39(npt-casette in pSB1C3(RFC10)).
Procedure:
- Ligation batch for F17+F40:
| volume | reagent |
| 1.63 µl | F17 (61.4 ng/µl, 2086 bp) |
| 1.88 µl | F40 (22.8 ng/µl, ~297 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 13.49 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F42+F38:
| volume | reagent |
| 1.36 µl | F42 (73.5 ng/µl, 2070 bp) |
| 3.42 µl | F38 (9.2 ng/µl, 217 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 12.22 µl | ddH2O |
| =20 µl | TOTAL |
- Ligation batch for F41+F39:
| volume | reagent |
| 1.11 µl | F41 (89.9 ng/µl, 2070 bp) |
| 13.74 µl | F39 (16.0 ng/µl, 1517 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 2.15 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
- Lid temperature = 37 °C
Thursday, May 30th
Gradient PCR of pActin (P143)
Investigator: Ingmar
Aim of the experiment: PCR of pActin
Procedure:
Operational sequence:
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq Reaction Standard Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O62 (pActin_for) |
| 1 µl | 10 µM Reverse Primer O63 (pActin_rev) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA P143 |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- PCR reaction mixture
| volume | reagent |
| 10 µl | 5x OneTaq GC Reaction Buffer |
| 1 µl | 10 mM dNTPs |
| 1 µl | 10 µM Forward Primer O62 (pActin_for) |
| 1 µl | 10 µM Reverse Primer O63 (pActin_rev) |
| 0.25 µL | OneTaq Hot Start DNA Polymerase (Finally: 1.25 units/50 µL) |
| 1 µl | Plasmid DNA P143 |
| 35.75 µL | ddH2O Water |
| =50 µL | TOTAL |
- Mix with pipette
- The PCR program was performed after following scheme:
| Initial denaturation | 94 °C | 30 s |
| 30 cycles | 94 °C | 30 s |
| 47 °C | 60 s | |
| 68 °C | 120 s | |
| Final extension | 68 °C | 5 min |
| Hold | 4 °C | infinite |
A temperature gradient was applied beginning at 47°C and ranging to 51°C. For each integer (47, 48, 49, 50 and 51°C) a single PCR batch was used.
- After PCR, the product was purified with QIAquick PCR Purification Kit, Qiagen.
Analytical gelelectrophoresis
Procedure:
- 4 µl of each PCR product were mixed seperately with 0.44 µl of DNA loading buffer (10x) and analytical gelelectrophoresis was performed at 1% agarose gel for 60 min at 90 V.
| 1 kbp ladder | F45 GC buffer | F46 GC buffer | F47 GC buffer | F47 GC buffer | F49 GC buffer | normal buffer | normal buffer | normal buffer | normal buffer | normal buffer | 1 kbp ladder |
| PCR products has expected length | PCR products has expected length | PCR products has expected length | PCR products has expected length | PCR products has expected length | PCR products has expected length, some byproducts | PCR products has expected length, some byproducts | PCR products has expected length, some byproducts | PCR products has expected length, some byproducts | PCR products has expected length, some byproducts |
Miniprep of overnight cultures of transformated E. coli XL1 blue of F17+F28 (FluA in pSB1C3 RFC25)
Investigator: Jeff, Flo
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue of F17+F28 (FluA in pSB1C3 RFC25).
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of ligation products F17+F28 (FluA in pSB1C3 RFC25)
Investigator: Flo, Jeff
Aim of the experiment: Analytical digestion and gelelectrophoresis of ligation products F17+F28 (FluA in pSB1C3 RFC25).
Procedure:
- Analytical digestion of ligation products of F17+F28 (FluA in pSB1C3 RFC25) with XbaI & AgeI.
| volume | reagent |
| 2.5 µl P150/P151/P152/P153 | |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | XbaI |
| 0.25 µl | AgeI-HF |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
| 1 kbp ladder DNA ladder# | Digestion of P150 with XbaI&AgeI-HF | Digestion of P151 with XbaI&AgeI-HF | Digestion of P152 with XbaI&AgeI-HF | Digestion of P153 with XbaI&AgeI-HF |
| as expected | as expected | as expected | as expected |
Preparative gelelectrophoresis of oligohybridization products F31 & F32
Investigator: Flo, Jeff
Aim of the experiment: Preparative gelelectrophoresis of oligohybridization products F31 & F32.
Procedure:
- 50 µl of the 1:10 dilution of F31 and F32 were mixed with 5 µl of DNA loading buffer (10x).
- Preparative gelelectrophoresis was performed 1.5 h on a 2% agarose gel at 90 V.
| 1 kbp ladder | F31 | F32 |
- Bands were extracted by QIAquick Gel Extraction Kit, QIAGEN.
- Gelpurified F31 and F32 were labelled as F43 and F44.
Transformation of E. coli XL1 blue with F17+F40 (PIF3 in pSB1C3(RFC25)), F42+F38 (t35S in pSB1C3(RFC10)) and F41+F39(npt-casette in pSB1C3(RFC10))
Investigator: Florian, Jeff
Aim of the experiment: Transformation of E. coli XL1 blue with F17+F40 (PIF3 in pSB1C3(RFC25)), F42+F38 (t35S in pSB1C3(RFC10)) and F41+F39(npt-casette in pSB1C3(RFC10)).
Procedure:
- CaCl2 competent E. coli XL1-Blue cells were put out from the stock in -80 °C freezer and were gently thawed on ice.
- 10 µl of DNA were added to 200 µl of competent cells and gently mixed.
- 30 min incubation on ice
- 5 min. heat shock at 37 °C
- Adding of 1 ml LB-medium to each tube.
- Incubation for 45 min at 37 °C in the 180 rpm cell-culture shaker.
- 100 µl of the cell suspension was plated on one chloramphenicol plate.
- The rest were centrifuged for 1 min at 13000 rpm and the supernatant was dicarded.
- The pellets were resuspended in 100 µl of LB-medium and these concentrated cell suspensions were plated again on a new chlorampenicol plates.
Ligation of F42+F43 and F17+F44
Investigator: Rosario
Picking of of E. coli XL1 blue transformed with EreB SDMIII (P149) and BBa_K801030 (SV40 NLS)
Investigator: Louise
Aim of the experiment: Picking of of E. coli XL1 blue transformed with EreB SDMIII (P149) and BBa_K801030 (SV40 NLS).
Procedure:
- Picked pipette tips was transferred into cell-culture tubes with air-permeable, sterile cover. Each tube contain 4 mL of LB-medium + 4 µL chloramphenicol(1000x).
- 5 colonies of EreB SDMIII (P149) and 2 colonies of BBa_K801030 (SV40 NLS) were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight.
Friday, May 31th
Miniprep of overnight cultures of transformated E. coli XL1 blue with P149 (QCIII of EreB) and BBa_K801030 (SV40 NLS)
Investigator: Louise, Jeff, Leonie, Katrin
Aim of the experiment: Miniprep of overnight cultures of transformated E. coli XL1 blue with P149 (QCIII of EreB) and BBa_K801030 (SV40 NLS)
Procedure:
- Miniprep was performed after manufacturer's protocol (QIAprep Miniprep, QIAGEN)
Analytical digestion and gelelectrophoresis of P149 (QCIII of EreB) with EcoRI-HF and PstI-HF
Investigator: Louise, Jeff, Leonie, Katrin
Aim of the experiment: Analytical digestion and gelelectrophoresis of P154-P158 (QCIII of EreB) with EcoRI-HF and PstI-HF to check if QCII was successful
Procedure:
- Analytical digestion of P154-P158 (QCIII of EreB) with EcoRI-HF and PstI-HF
| volume | reagent |
| 2.5 µl | P149-P158/P133 (control before QCIII) |
| 2 µl | CutSmart Buffer (10x) |
| 0.25 µl | EcoRI-HF |
| 0.25 µl | PstI-HF |
| 15 µl | ddH2O |
| =20 µl | TOTAL |
- Incubation for 90 min at 37 °C.
- Analytical gelelectrophoresis was performed at 90 V for 60 min.
[[File:|500px]]
| 1 kbp ladder DNA ladder# | Digestion of P154 with EcoRI-HF and PstI-HF | Digestion of P155 with EcoRI-HF and PstI-HF | Digestion of P156 with EcoRI-HF and PstI-HF | Digestion of P157 with EcoRI-HF and PstI-HF | Digestion of P158 with EcoRI-HF and PstI-HF | Digestion of P133 with EcoRI-HF and PstI-HF |
| as expected | as expected | as expected | as expected | as expected | as expected |
Sequencing of P157 and P160
Investigators: Louise, Jeff, Leonie, Katrin
Aim of the experiment: Sequencing of QCIII product (EreB) and BBa_K801030 (SV40 NLS)
Procedure: The DNA was prepared for sequencing according to the Eurofins SmartSeq Kit protocol (15 µl of plasmid DNA (50 - 100 ng) and 2 µl sequencing primer).
The different genes we sequenced received the following barcodes:
| Label | Name | Barcode1 | Barcode2 |
|---|---|---|---|
| P157 | Miniprep of transformation of QCIII product of P149 (QC - SDMIII - of EreB to remove forbidden PstI) | FR01002291 (VR) | FR01002292 (VF2) |
| P160 | Miniprep of transformation of BBa_K801030 (SV40 NLS) | FR01002289 (VR) | FR01002290 (VF2) |
Ligation of F29+F30
Investigator: Leonie, Katrin
Aim of the experiment: Ligation of F29+F30
Procedure:
- Ligation batch
| volume | reagent |
| 4.22 µl | F29 (23.7 ng/µl, 2110 bp)) |
| 10.48 µl | F30 (9,5 ng/µl, about 700 bp) |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 2.3 µl | ddH2O |
| =20 µl | TOTAL |
Ligation was performed at 37 °C overnight
Quick Change mutagenesis to remove forbidden restriction sites - SDMIII - of P124 (Laccase)
Investigator: Louise, Leonie, Katrin
Aim of the experiment: Removal of forbidden restiction site from P124 (Laccase).
Procedure: Quickchange - PCR
Reaction batch
| volume | reagent | Additional information |
| 2.5 µl | 10x Pfu Ultra II buffer | |
| 1 µl | Plasmid template (P124 - 1:10 dilution) | need to get between 5 and 50 ng/µl |
| 1 µl | 1:10 dilution of O30 (for P124, 10 µM) | |
| 1 µl | 1:10 dilution of O31 (for P124, 10 µM) | |
| 18 µl | ddH2O | |
| 1 µl | dNTP mix 50mM | |
| 0.5 µl | Pfu Ultra II DNA polymerase (2.5 U / µl) |
PCR cycling parameters - P124
| Segment | Cycles | Temperature | Time |
| 1 | 1 | 95 °C | 30 s |
| 2 | 19 | 95 °C | 30 s |
| 52 °C | 1 min | ||
| 68 °C | 4 min | ||
| 4 °C | hold |
After the PCR; 1 µl of Dpn1 was added and the reaction mixture was incubated at 37°C for one hour
Preparative digestion of pActin(F46) with XbaI & PstI
Investigator: Ingmar
Aim of the experiment: Preparative digestion of pActin(F46) in order to ligat this fragment in pSB1C3 later on.
Procedure:
- Batch for preparative digestion of pActint(F46) with XbaI and PstI
| volume | reagent |
| 25 µl | F46 PCR product of pActin |
| 5 µl | CutSmart Buffer |
| 1 µl | XbaI (20 U/µl) |
| 1 µl | PstI (20 U/µl) |
| 18 µl | ddH2O |
| =50 µl | TOTAL |
- Incubation for 3 h at 37 °C.
- 5 µl of DNA loading buffer (10x) were added to the 50 µl reaction batchesand loaded on a 1% agarose gel for preparative gelelectrophoresis.
- Preparative gelelectrophoresis was performed at 90 V for 90 min.
- The band was extracted by QIAquick Gel Extraction Kit, QIAGEN. The resulting fragment was named: F50
Ligation of F42+F50 (pActin in pSB1C3)
Investigator: Ingmar
Aim of the experiment: Ligation of F42+F50 (pActin in pSB1C3(RFC25).
Procedure:
- Ligation batch
| volume | reagent |
| 1.37 µl | F42 |
| 13.13 µl | F50 |
| 2 µl | T4 ligase buffer (10x) |
| 1 µl | T4 ligase (1 U/µl) |
| 2.5 µl | ddH2O |
| =20 µl | TOTAL |
- Negative control was also prepared. Instead of the insert, the same volume of ddH2O was added to the reaction batch.
- Cycled ligation has been performed after following protocol in a thermocycler:
| 99 cycles | 12 °C | 60 s |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| 12 °C | 60 s | |
| 22 °C | 60 s | |
| Hold | 16 °C | infinite |
- Lid temperature = 37 °C
Picking of of E. coli XL1 blue transformed with F42+F38 (t35S), F17+F40 (Pif 3) and F41+F39 (npt)
Investigator: Ingmar
Aim of the experiment: Picking of of E. coli XL1 blue transformed with F42+F38 (t35S), F17+F40 (Pif 3) and F41+F39 (npt).
Procedure:
- The picked colonies were transferred into cell-culture tubes with air-permeable, sterile cover containing 5 mL of LB-medium + 5 µL chloramphenicol(1000x).
- 3 colonies of each agar plate were picked.
- These tubes were transferred in a cell culture shaker at 37 °C and were incubated overnight.
Saturday, june 1st
Transformation
Investigator: Leonie, Katrin
Miniprep
Investigator: Leonie, Katrin
Analytical digestion
Investigator: Leonie, Katrin
 "
"