04/09/13
From 2013.igem.org
(Difference between revisions)
(→Ligation of the TOD genes (X, F and ToBG) to the pSB1C3 backbone) |
|||
| Line 77: | Line 77: | ||
|Samples || A (TodX) ||C (TodF) ||E (ToBG)||RFP-sample 2 (positive control)||Background control | |Samples || A (TodX) ||C (TodF) ||E (ToBG)||RFP-sample 2 (positive control)||Background control | ||
|- | |- | ||
| - | |2X Rapid ligation buffer|| 5|| 5|| 5|| 5|| 5 | + | |2X Rapid ligation buffer (ul)|| 5|| 5|| 5|| 5|| 5 |
|- | |- | ||
| - | |vector (pSB1C3-sample1)|| 1.6|| 1.6|| 1.6|| 1.6|| 1.6 | + | |vector (pSB1C3-sample1) (ul)|| 1.6|| 1.6|| 1.6|| 1.6|| 1.6 |
|- | |- | ||
| - | |T4 DNA ligase|| 1|| 1|| 1|| 1|| 1 | + | |T4 DNA ligase (ul) || 1|| 1|| 1|| 1|| 1 |
|- | |- | ||
| - | |PCR product|| 2.3|| 1.4|| 3.1|| 3.6|| - | + | |PCR product (ul)|| 2.3|| 1.4|| 3.1|| 3.6|| - |
|- | |- | ||
|Deionized water for the final volume of 12 ul|| 2.1|| 3|| 1.3|| 0.8|| 4.4 | |Deionized water for the final volume of 12 ul|| 2.1|| 3|| 1.3|| 0.8|| 4.4 | ||
|} | |} | ||
| + | |||
| + | The reaction were incubated overnight at 4C for the maximum number of transformants | ||
Revision as of 09:39, 5 September 2013
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Contents |
Transformation results
Gel Electrophoresis
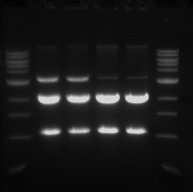
- The double digest (pSB1C3) from yesterday was run on a 1% gel.
- This was done in order to confirm that the digest worked and also to purify the pSB1C3 vector.
- From left to right
- Sample 1.1 - track 2
- Sample 1.2 - track 3
- Sample 2.1 - track 4
- Sample 2.2 - track 5
- The 2 Kb bands are the pSB1C3 backbone
- The 1 Kb bands are the RFP biobrick
The two higher bands in tracks 2 and 3 are plasmids that were not digested.
Gel purification of the pSB1C3 backbone and RFP biobrick
- The Zymoclean Gel DNA recovery Kit was used to perform the purification and its protocol was followed.
- However the elution step was changed to 12ul of elution buffer, and this step was repeated.
Nanodrop of the samples
| Sample | Volume | Concentration ng/ul | 260/280 | 260/230 |
| pSB1C3 1 | 19.8 | 31.9 | 1.83 | 1.75 |
| pSB1C3 2 | 20 | 34 | 1.67 | 0.92 |
| RFP 1 | 20 | 28.8 | 1.57 | 0.71 |
| RFP 2 | 18.8 | 22.2 | 1.79 | 1.84 |
Double digest of TOD genes PCR products
Samples A (TodX), C (TodF) and E (ToBG) were digested with the enzymes XbaI and SpeI.
- Protocol
| Samples | A | C | E |
| Volume (ul) | 20 | 23.5 | 21 |
| SpeI (ul) | 1 | 1 | 1 |
| XbaI | 0.5 | 0.5 | 0.5 |
| Cutsmart Buffer | 6 | 6 | 6 |
| 5mTris Hcl (ul) | 32.5 | 29 | 31.5 |
- The samples were incubated at 37C for 90 minutes
- Heat kill the enzymes by incubating the samples at 80C for 20 minutes
Ligation of the TOD genes (X, F and ToBG) to the pSB1C3 backbone
The pGEM-T Vector System was used, However changes were made to the protocol. The promega biomath caculator (www.promega.com/biomath) was used to work out the ratio of insert to vector. The DNA concentration of the TOD genes can be found at Lab work 28/09/2013. The DNA concentration of the pSB1C3 and the RFP biobrick can be found above. The amount of vector DNA was added at 50ng.
Protocol
| Samples | A (TodX) | C (TodF) | E (ToBG) | RFP-sample 2 (positive control) | Background control |
| 2X Rapid ligation buffer (ul) | 5 | 5 | 5 | 5 | 5 |
| vector (pSB1C3-sample1) (ul) | 1.6 | 1.6 | 1.6 | 1.6 | 1.6 |
| T4 DNA ligase (ul) | 1 | 1 | 1 | 1 | 1 |
| PCR product (ul) | 2.3 | 1.4 | 3.1 | 3.6 | - |
| Deionized water for the final volume of 12 ul | 2.1 | 3 | 1.3 | 0.8 | 4.4 |
The reaction were incubated overnight at 4C for the maximum number of transformants
 "
"