Team:BYU Provo/Notebook/LargePhage/Summerexp/Period1/Dailylog
From 2013.igem.org
(→7/10/13) |
|||
| (24 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{TeamBYUProvo}} | {{TeamBYUProvo}} | ||
| + | <br> | ||
| - | = | + | {| width="100%" |
| - | === | + | | colspan="3" | <font color="#333399" size="5" font face="Calibri"> |
| - | + | : '''Large Phage July - August Notebook: September 1 - September 15 Daily Log'''</font> | |
| - | + | <br> | |
| - | + | <br> | |
| - | == | + | |- valign="top" |
| - | + | | style="width: 22%; background-color: transparent;"| | |
| - | + | <font color="#333399" size="3" font face="Calibri"> | |
| - | < | + | |
| - | + | ||
| + | <font size = "4"> | ||
| - | + | : <u> '''Large Phage''' </u> </font> | |
| - | < | + | |
| - | < | + | |
| - | + | : [[Team:BYU Provo/Notebook/LargePhage/Winterexp|March-April]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | : [[Team:BYU Provo/Notebook/LargePhage/Springexp|May-June]] | |
| - | + | : [[Team:BYU Provo/Notebook/LargePhage/Summerexp|July-August]] | |
| - | + | : [[Team:BYU Provo/Notebook/LargePhage/Fallexp|September-October]] | |
| - | [[ | + | |
| - | + | </font> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | | style="width: 82%; background-color: transparent;"| | |
| - | + | ||
| - | + | ||
| - | == | + | <font face="Calibri" size="3"> |
| - | + | ||
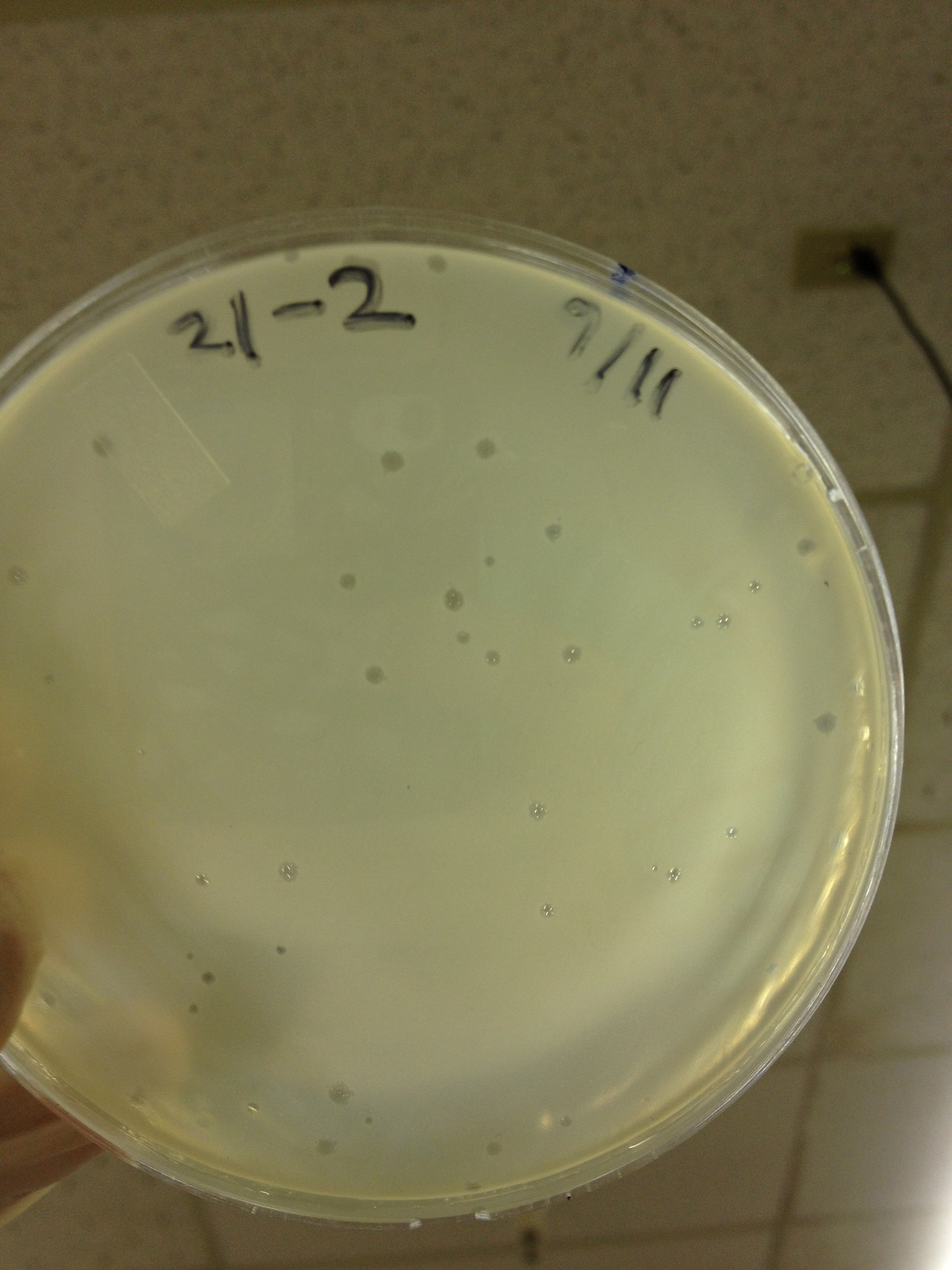
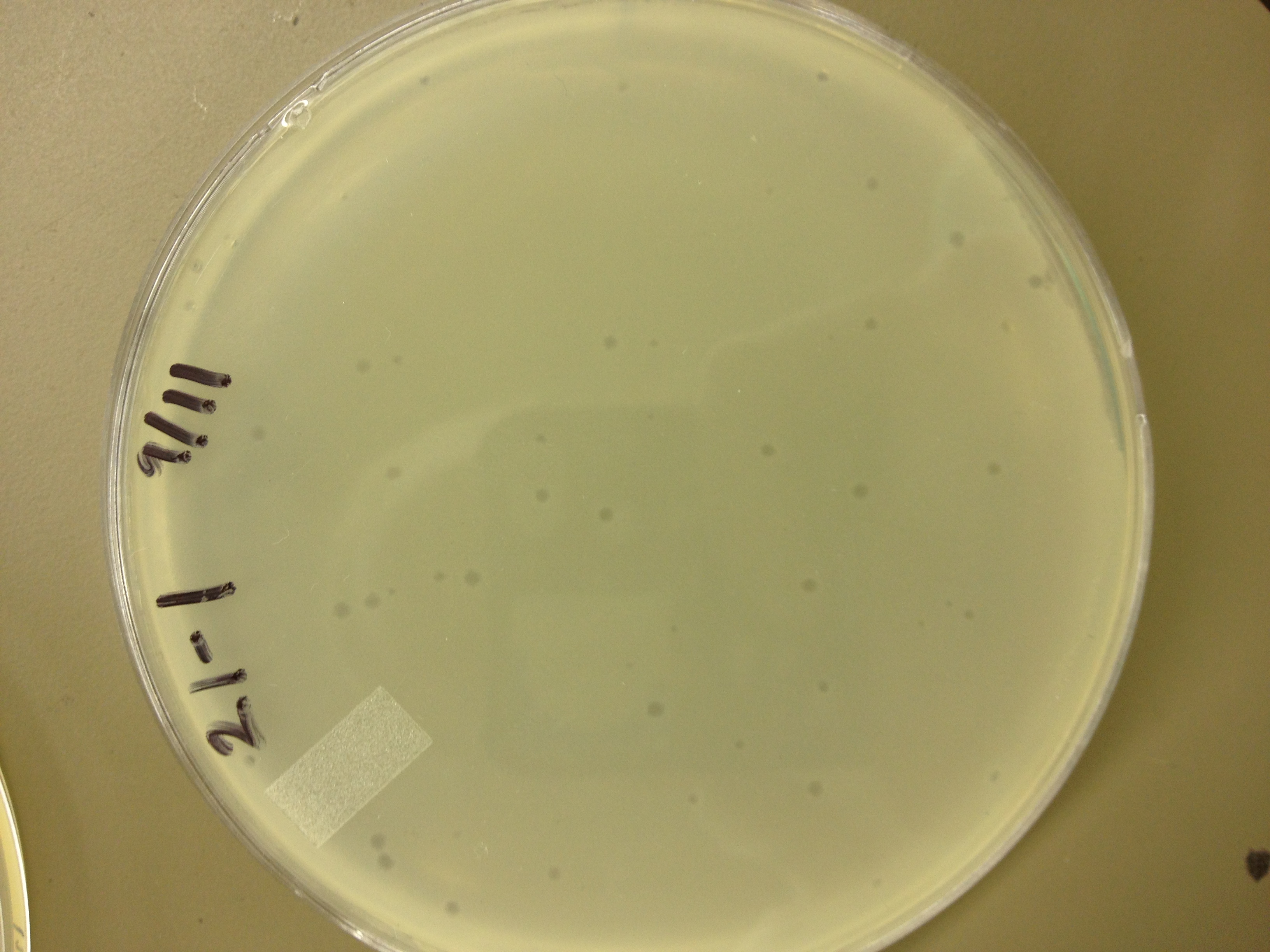
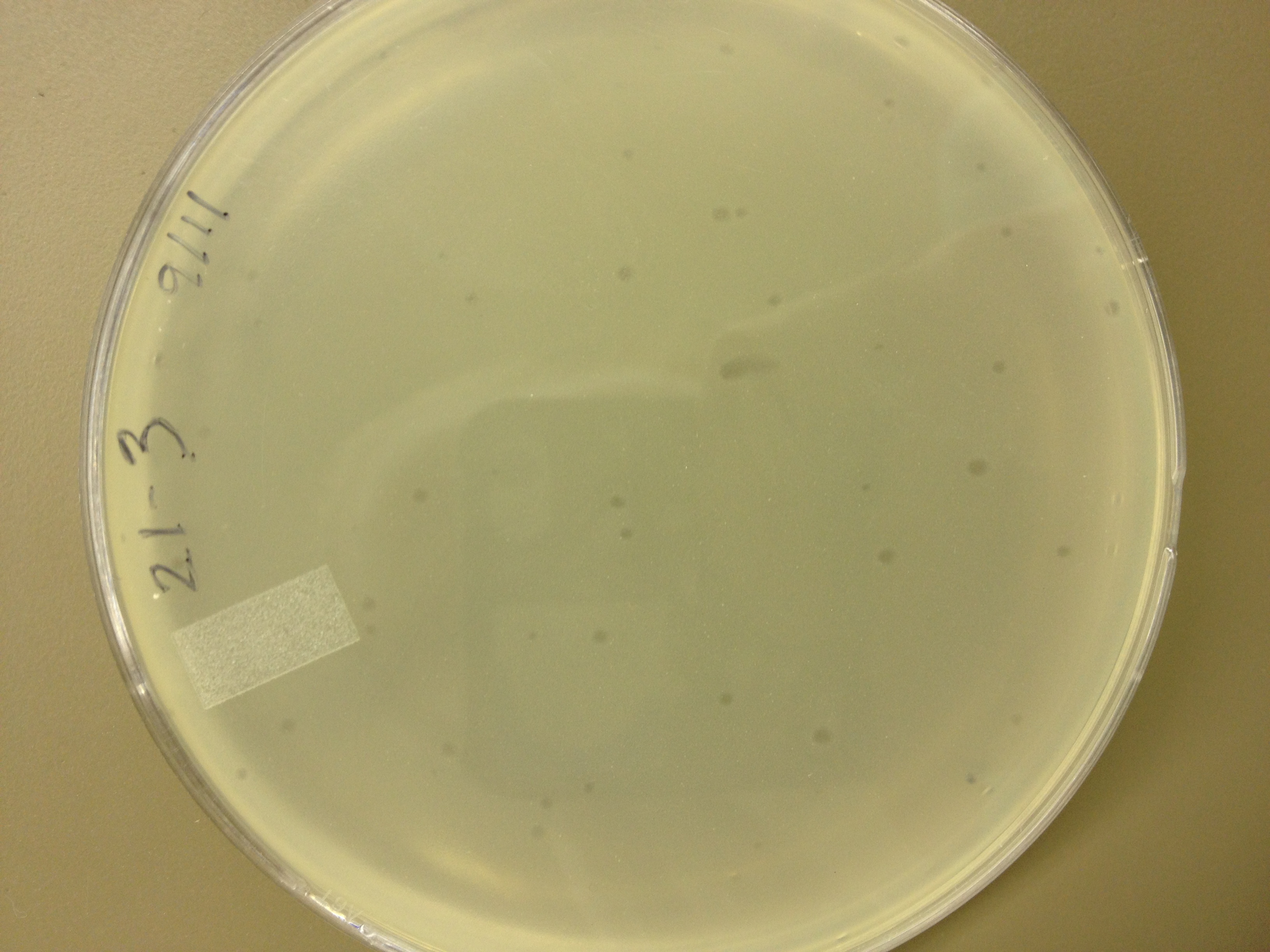
| - | = | + | <font size="4"> '''9/2/13''' </font> |
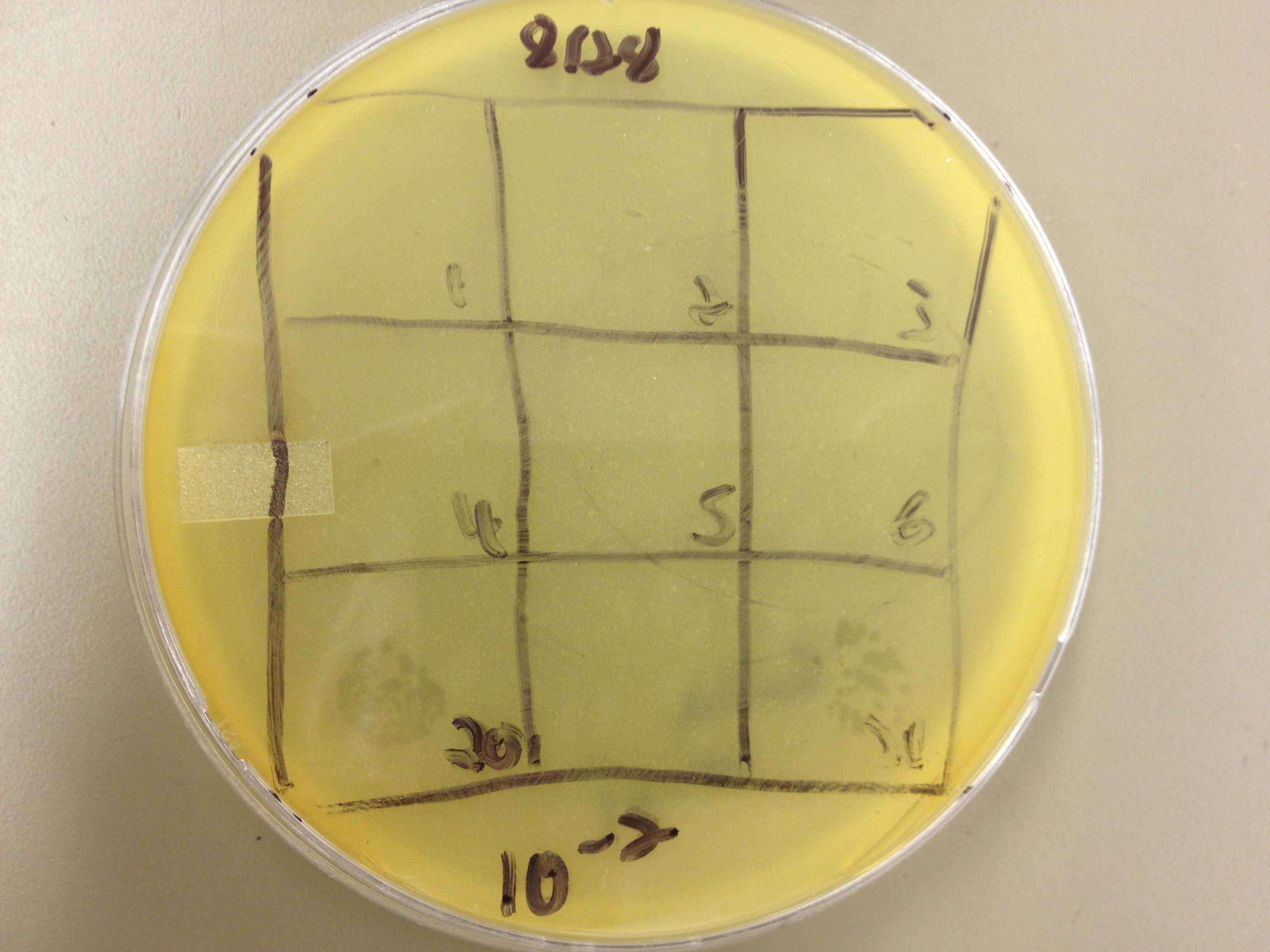
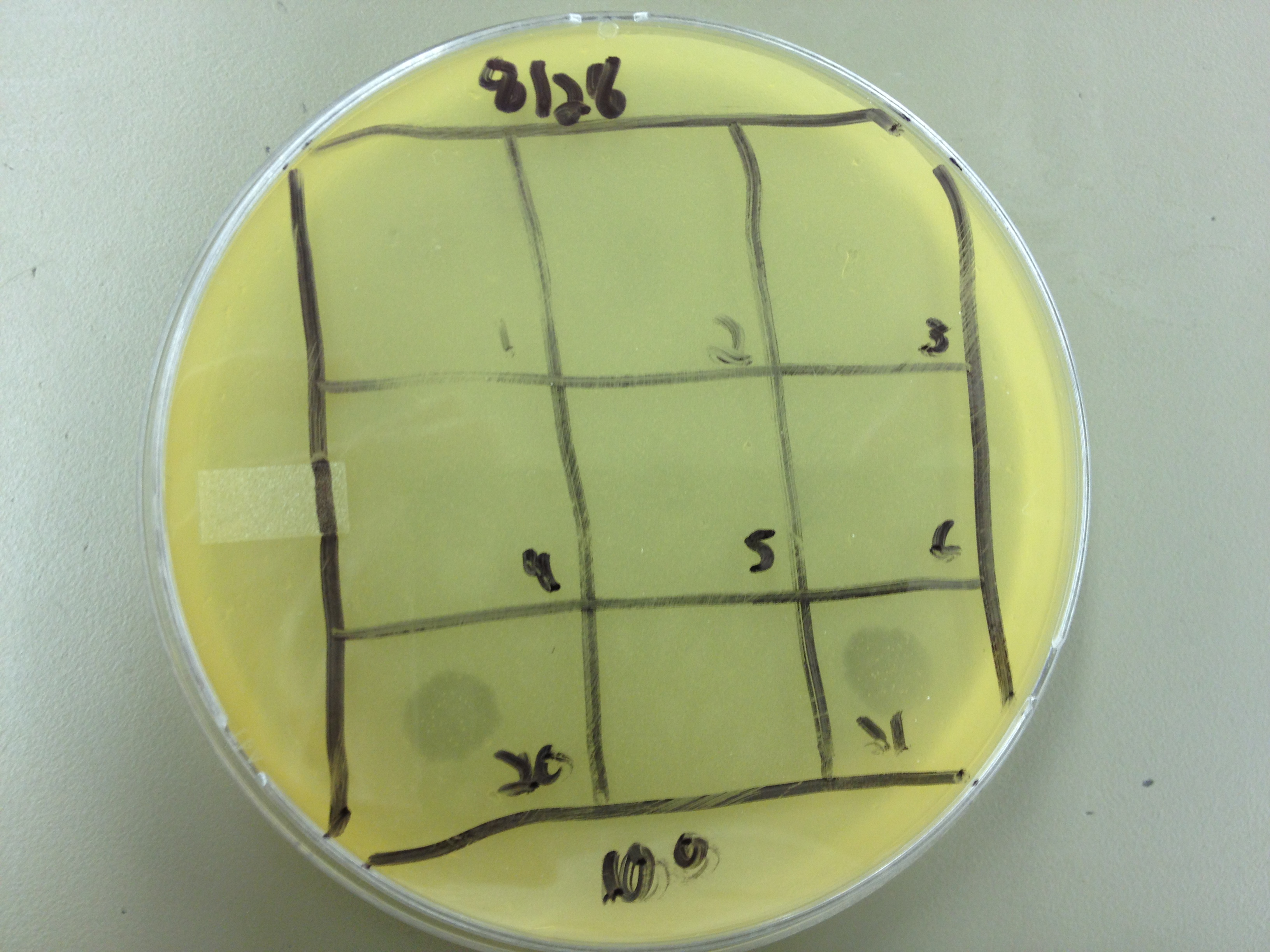
| - | + | Results from 8/28/13: Fractions 20 and 21 produced clearage on 10^0, and many small plaques at 10^-2. #4 had one plaque on 10^-2, and #1 had one plaque on 10^0. | |
| - | + | Today we infected 0.5 mL of E. coli with: 100 uL of #1 10^0, 100 uL of #4 10^0, 10 uL of #20 10^-2, 10 uL of #21 10^-2, and for controls we did 10 uL of T4 wild type 10^-6 and 1 uL of T4 10^-7. We also plated a lawn of E. coli as a control. | |
| - | |||
| - | + | [[File:September1-0.jpg|400px]] | |
| - | + | [[File:September1-1.jpg|400px]] | |
| - | + | BDM KW | |
| - | + | <br> | |
| - | = | + | <font size="4"> '''9/6/13''' </font> |
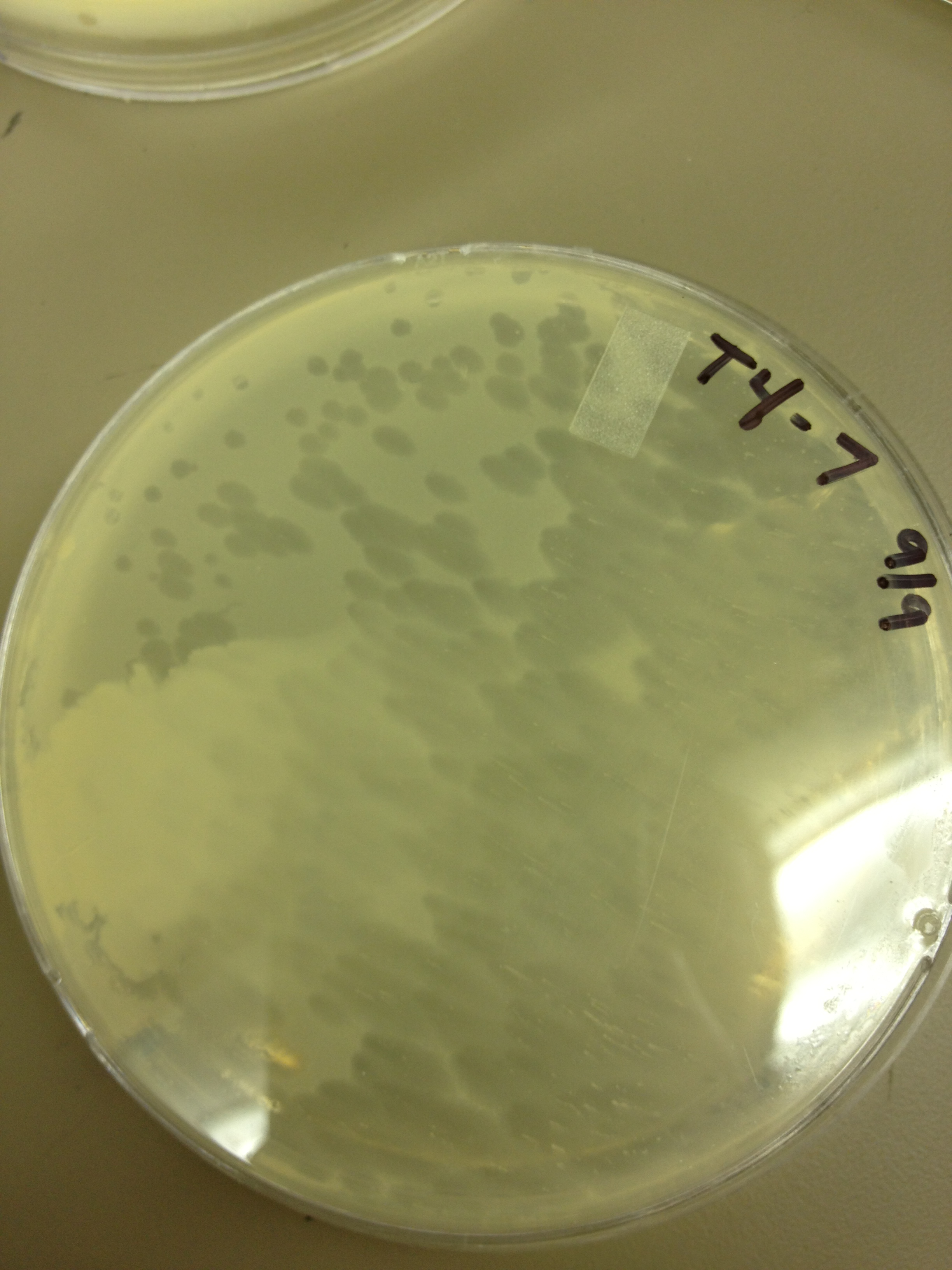
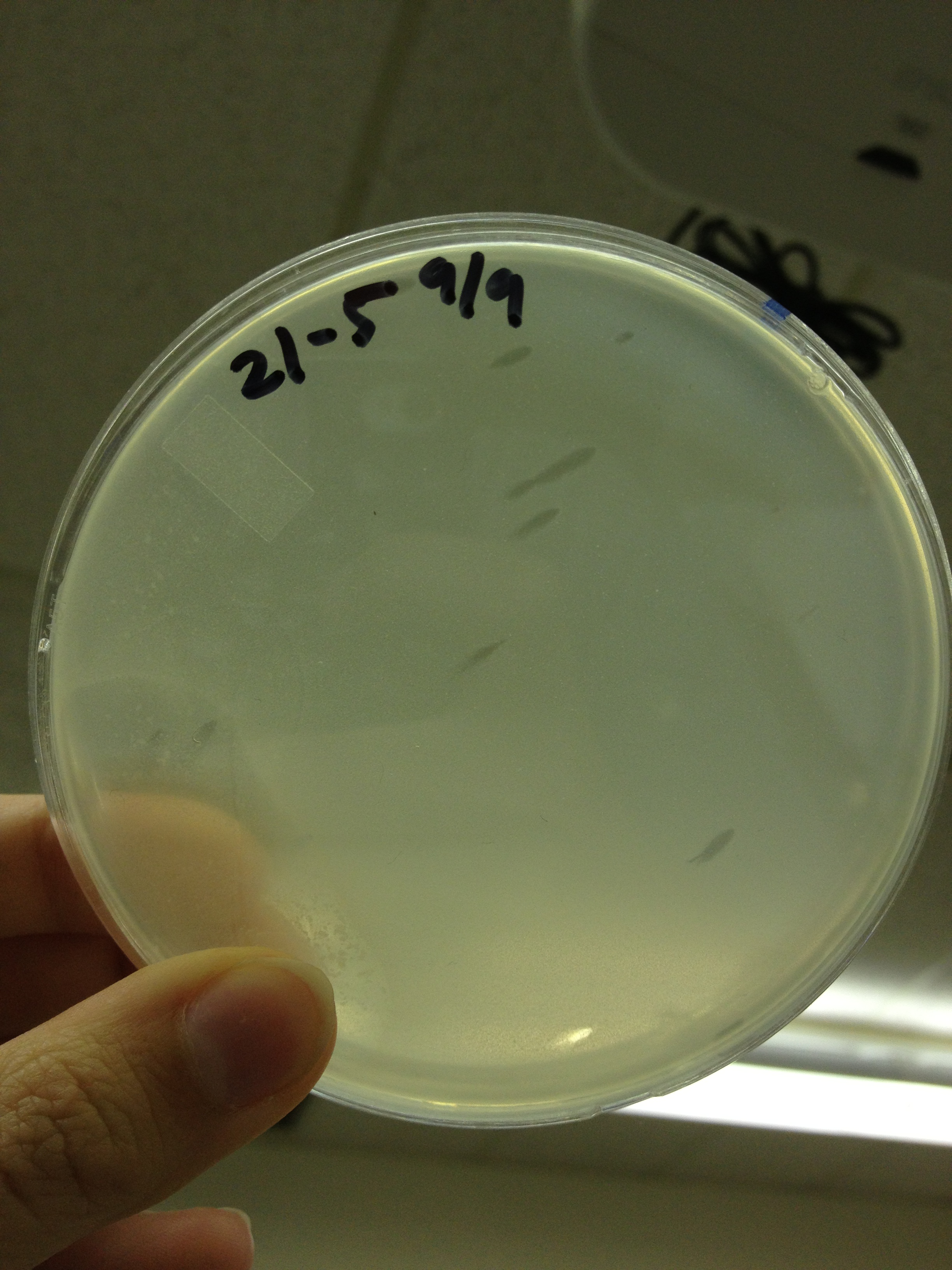
| + | Results from 9/2: | ||
| - | |||
| - | + | [[File:September2-0.jpg|400px]] | |
| - | + | [[File:September2-1.jpg|400px]] | |
| - | + | [[File:September2-2.jpg|400px]] | |
| - | + | ||
| - | + | [[File:September2-3.jpg|400px]] | |
| - | |||
| - | + | In general, the plaques for the mutagenized phage seem to be smaller than the pre-mutagenized phage. From this it would appear that we have large phage! But, it's important to make sure that we have a stable phage, so from here we will be picking the small plaques from the mutant plate and plating them out so we can observe if there is a steady result of small plaques. | |
| + | BDM,KW | ||
| - | + | <br> | |
| - | = | + | <font size="4"> '''9/9/13''' </font> |
| - | + | Today we picked plaques from phages found in the 20 and 21 fractions of the Cesium Chloride gradient that had the smallest plaques. We plated them out with .075 agarose in hopes that the plaques would be bigger on the whole, so differences would be more easily seen. | |
| - | + | The results were somewhat disappointing, it appeared that each plaques was smeared either because we allowed the phage to infect e.coli too long before we plated them, because the concentration was not high enough for the agarose to solidify, or a combination of the two. | |
| - | |||
| - | + | [[File:September9.jpg|400px]] | |
| - | + | [[File:September9-1.jpg|400px]] | |
| - | + | [[File:September9-2.jpg|400px]] | |
| - | = | + | [[File:September9-3.jpg|400px]] |
| + | |||
| + | [[File:September9-4.jpg|400px]] | ||
| + | |||
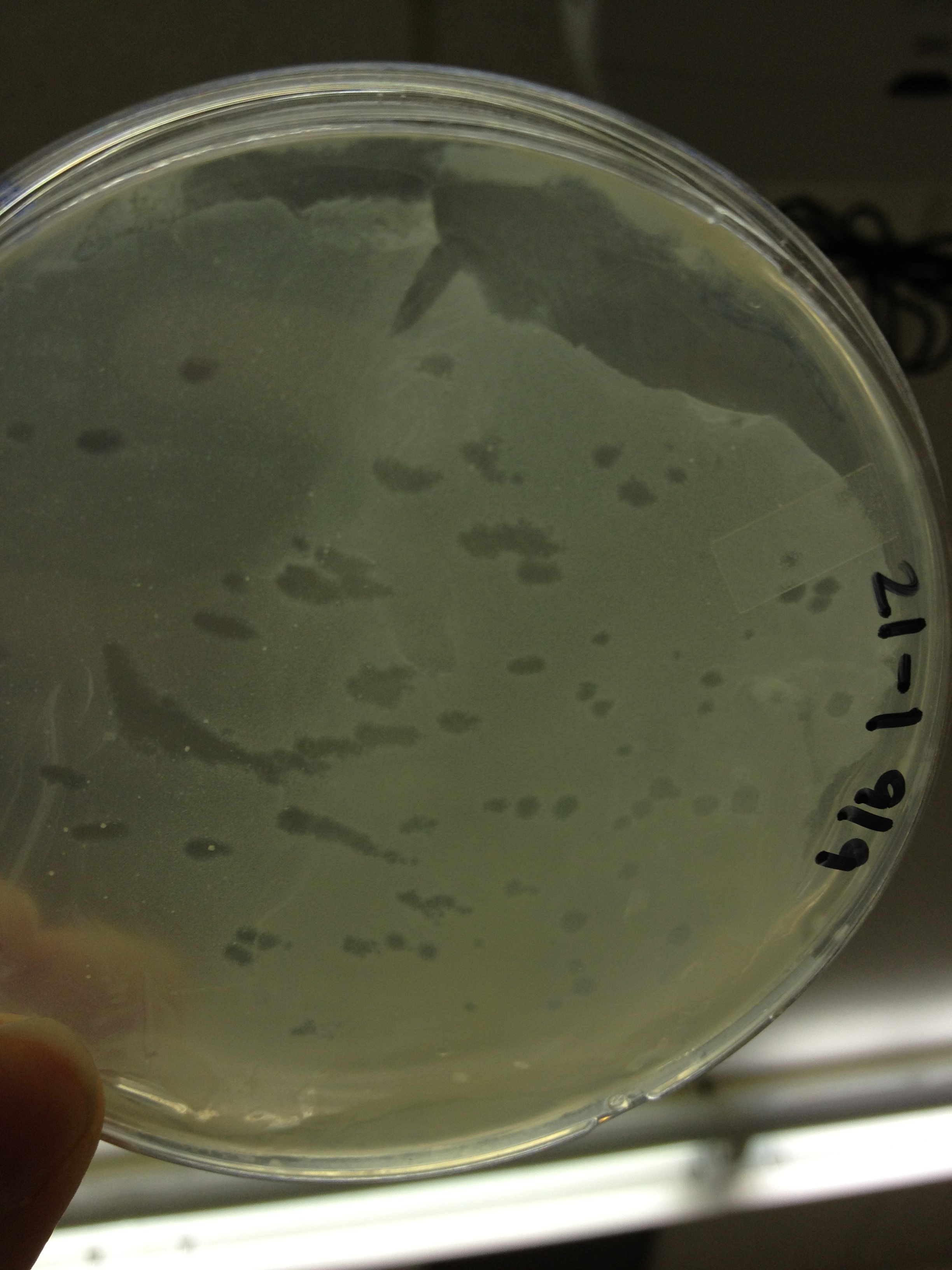
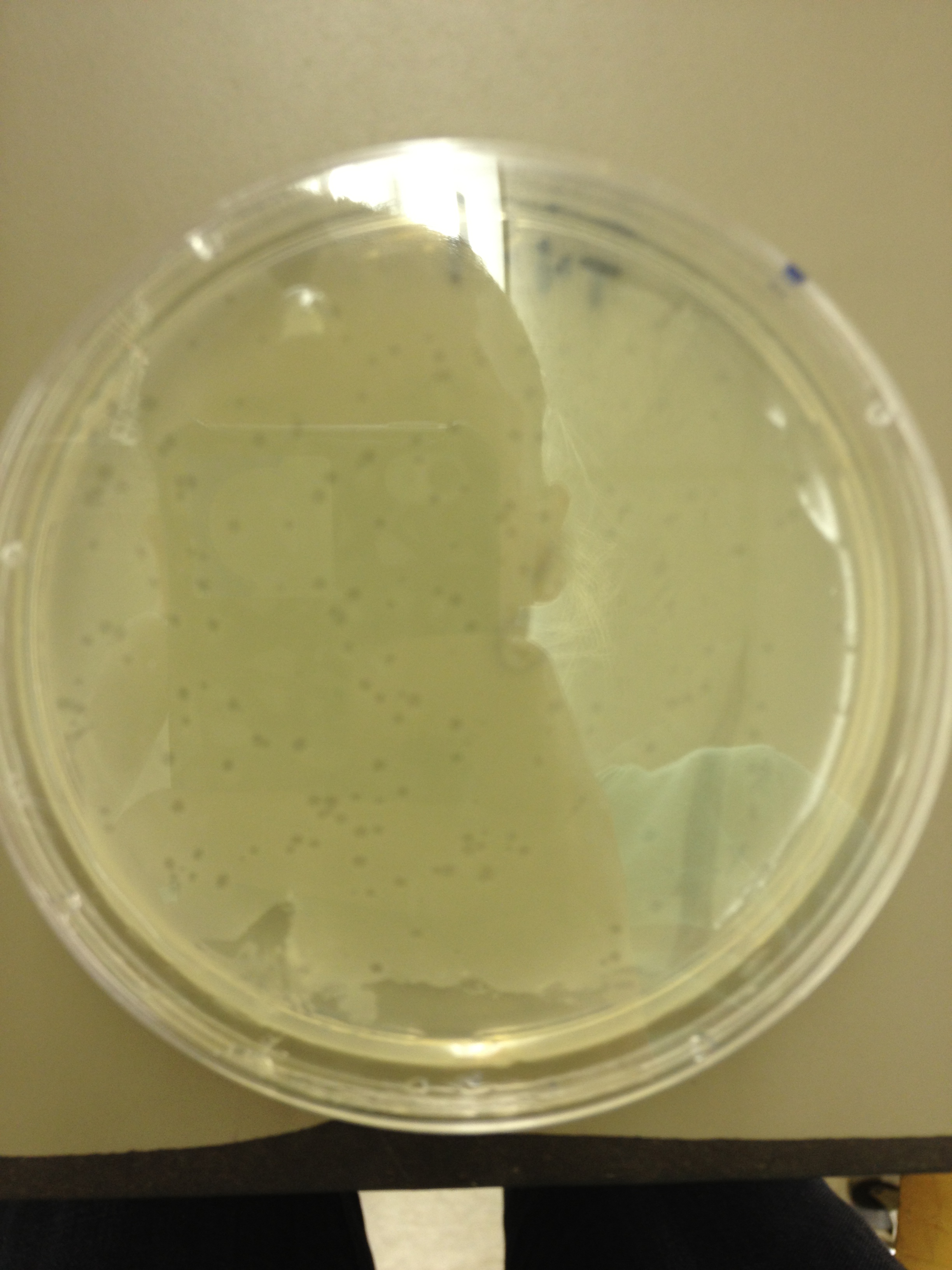
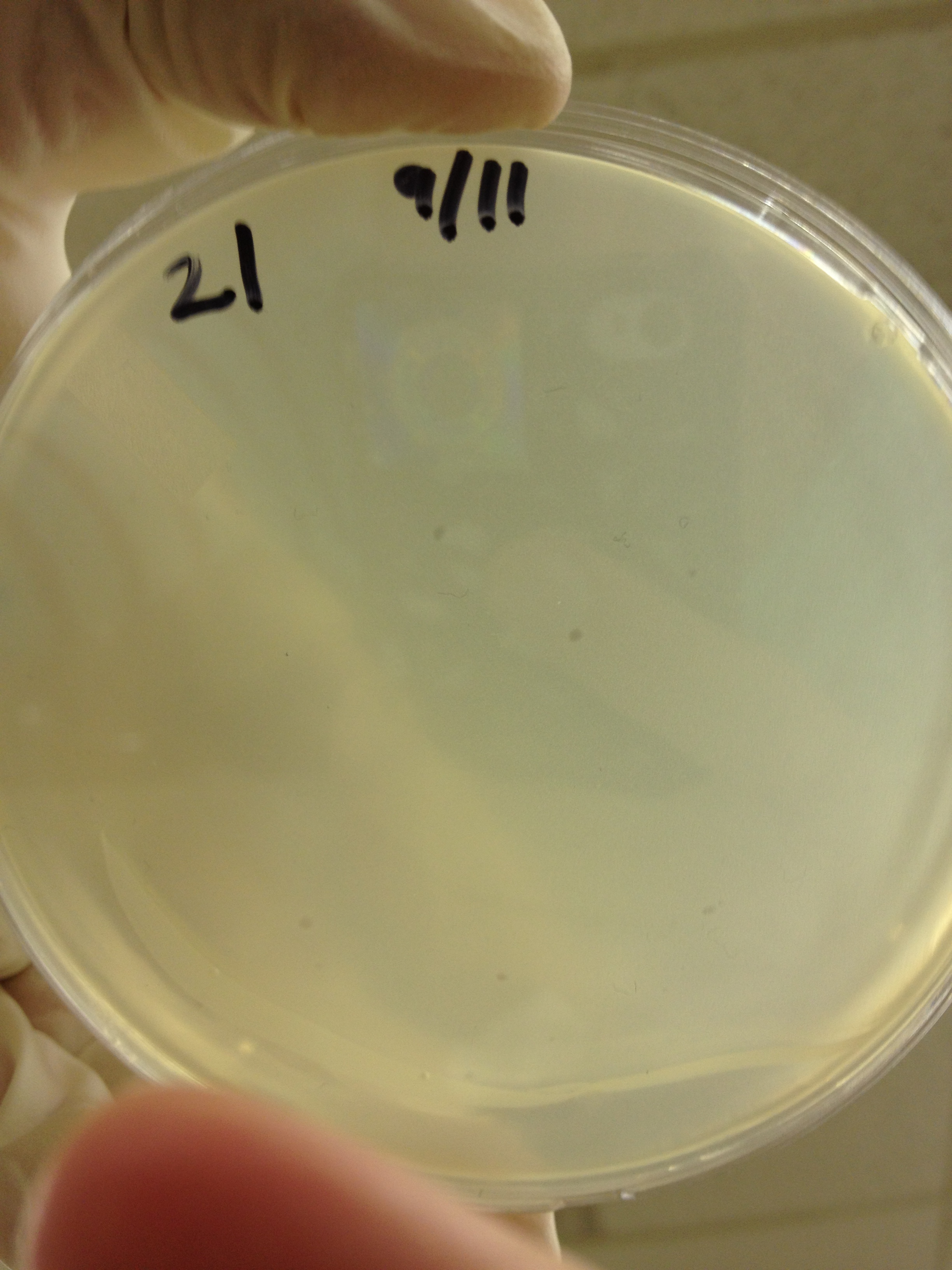
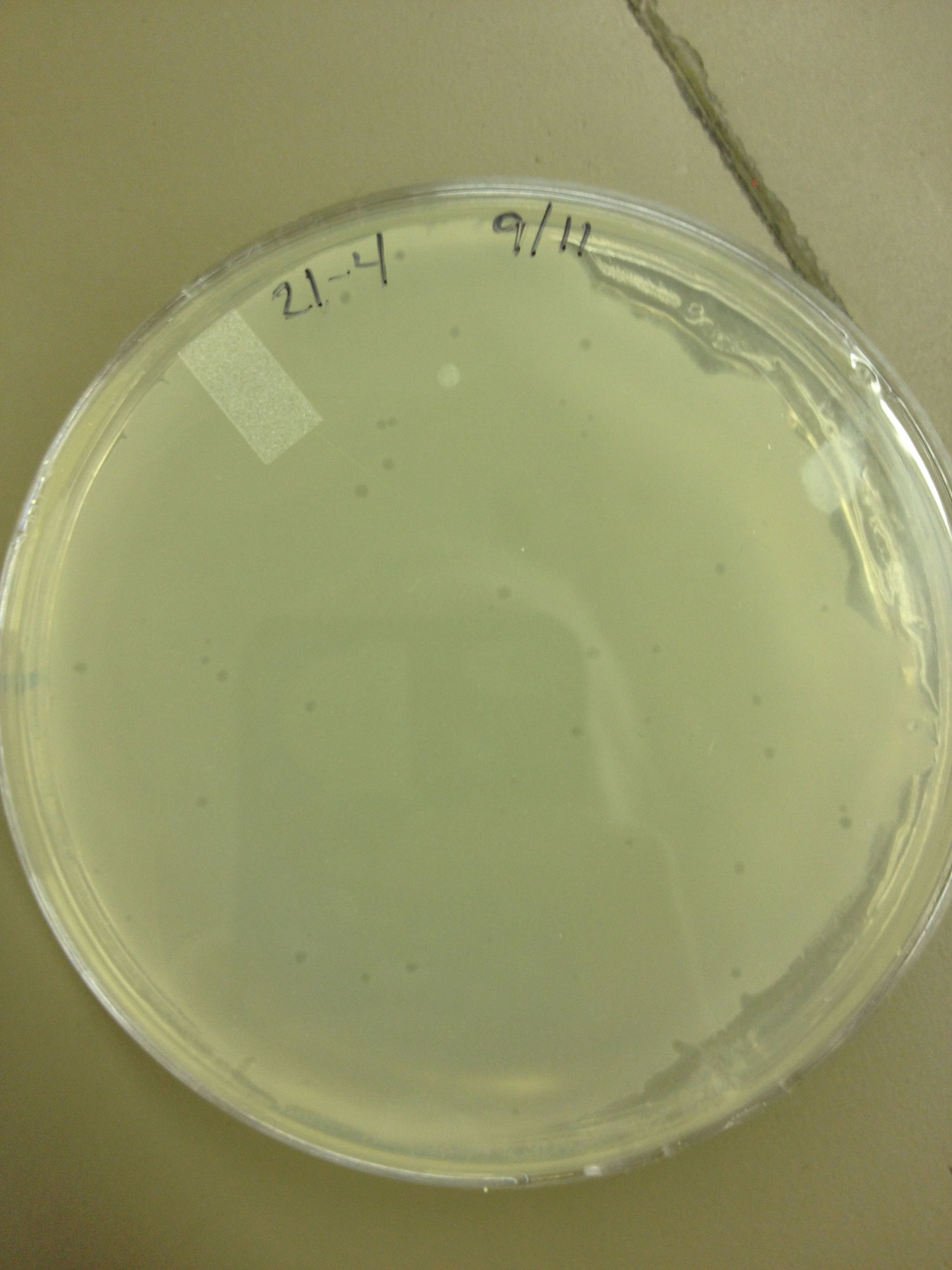
| + | <font size="4"> '''9/11/13''' </font> | ||
| + | |||
| + | |||
| + | [[File:September11.jpg|400px]] | ||
| + | |||
| + | [[File:September11-1.jpg|400px]] | ||
| + | |||
| + | [[File:September11-2.jpg|400px]] | ||
| + | |||
| + | [[File:September11-3.jpg|400px]] | ||
| + | |||
| + | [[File:September11-4.jpg|400px]] | ||
| + | |||
| + | [[File:September11-5.jpg|400px]] | ||
| + | |||
| + | [[File:September11-6.jpg|400px]] | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <font size="4"> '''9/13/13''' </font> | ||
| + | |||
| + | We picked plaques and put them in eppendorf tubes numbered: | ||
| + | #1: T4 control phage | ||
| + | #2: 2 on 20-7 | ||
| + | #3: 3 on 20-7 | ||
| + | #4: 4 on 20-7 | ||
| + | |||
| + | We grew liquid cultures of #1 and #2 (10 uL in .5 mL of E. coli B) for 3 hours. We then added 100 uL of chloroform and pelleted the bacteria. | ||
| + | |||
| + | We prepared seven EM grids (300 mesh). Each grid was prepared by adding 10 uL of sample to the grid for 1 minute, wicking off the droplet with filter paper, adding 10 uL of 2% phosphotungstic acid for 1 minute, wicking off the droplet, and placing back in the grid box. | ||
| + | Grid label Sample description | ||
| + | |||
| + | These next two samples were prepared by inserting a 23.5 gauge needle in a plaque on a plate and rinsing it in 50 uL of LB broth. | ||
| + | B7 - T4 fraction 21 plaque 7-1 | ||
| + | B8 - T4 fraction 21 plaque 7-2 | ||
| + | These three samples were prepared by incubating E. coli and T4 together to try to get pictures of both. | ||
| + | B6 - T4 + E. coli incubated together for 2 minutes | ||
| + | B9?- T4 incubated with E. coli for 10 mins | ||
| + | C7 - T4 incubated with E. coli for 40 mins | ||
| + | These two samples were prepared by trying to propagate 10 uL of the samples used in B7 and B8 using a liquid culture technique as described above. | ||
| + | B10 - T4 fraction 21 plaque 7-1 liquid culture | ||
| + | C6 - T4 fraction 21 plaque 7-2 liquid culture | ||
| + | |||
| + | We imaged all grids except B10. Also, B9 may be mislabeled as some of the grids fell out of the box as the technician was loading the grid into the machine. | ||
| + | |||
| + | |||
| + | KW BDM | ||
| + | |||
| + | <br> | ||
{{TeamBYUProvoFooter}} | {{TeamBYUProvoFooter}} | ||
Latest revision as of 17:53, 27 September 2013
| ||
|
|
9/2/13 Results from 8/28/13: Fractions 20 and 21 produced clearage on 10^0, and many small plaques at 10^-2. #4 had one plaque on 10^-2, and #1 had one plaque on 10^0. Today we infected 0.5 mL of E. coli with: 100 uL of #1 10^0, 100 uL of #4 10^0, 10 uL of #20 10^-2, 10 uL of #21 10^-2, and for controls we did 10 uL of T4 wild type 10^-6 and 1 uL of T4 10^-7. We also plated a lawn of E. coli as a control. BDM KW
9/6/13 Results from 9/2:
9/9/13 Today we picked plaques from phages found in the 20 and 21 fractions of the Cesium Chloride gradient that had the smallest plaques. We plated them out with .075 agarose in hopes that the plaques would be bigger on the whole, so differences would be more easily seen. The results were somewhat disappointing, it appeared that each plaques was smeared either because we allowed the phage to infect e.coli too long before we plated them, because the concentration was not high enough for the agarose to solidify, or a combination of the two. 9/11/13
9/13/13 We picked plaques and put them in eppendorf tubes numbered:
We grew liquid cultures of #1 and #2 (10 uL in .5 mL of E. coli B) for 3 hours. We then added 100 uL of chloroform and pelleted the bacteria. We prepared seven EM grids (300 mesh). Each grid was prepared by adding 10 uL of sample to the grid for 1 minute, wicking off the droplet with filter paper, adding 10 uL of 2% phosphotungstic acid for 1 minute, wicking off the droplet, and placing back in the grid box. Grid label Sample description These next two samples were prepared by inserting a 23.5 gauge needle in a plaque on a plate and rinsing it in 50 uL of LB broth. B7 - T4 fraction 21 plaque 7-1 B8 - T4 fraction 21 plaque 7-2 These three samples were prepared by incubating E. coli and T4 together to try to get pictures of both. B6 - T4 + E. coli incubated together for 2 minutes B9?- T4 incubated with E. coli for 10 mins C7 - T4 incubated with E. coli for 40 mins These two samples were prepared by trying to propagate 10 uL of the samples used in B7 and B8 using a liquid culture technique as described above. B10 - T4 fraction 21 plaque 7-1 liquid culture C6 - T4 fraction 21 plaque 7-2 liquid culture We imaged all grids except B10. Also, B9 may be mislabeled as some of the grids fell out of the box as the technician was loading the grid into the machine.
|
|
 "
"