Team:Chiba/Parts
From 2013.igem.org

Parts
Golden Gate
1. Introduction
Gentic switch such as pBAD/araC system is very useful for overexpression of given genes.
In order to place the various open reading frames with its RBS under the pBAD/AraC system, we improved BBa_I746908 to insert BsaI site in both sides of sfgfp gene. Owing to this improvement, we were able to use Golden Gate Method (ref. Carola Engler et al) as below:
⑴ まずベクターをBsaIで切る
⑵ インサートはbsaIとRBSつけてPCR-up
⑶インサートとベクター混ぜて、ダイジェストとらいげーとをいっぺんにやる
This method is designed that BsaI site doesn't remain on the vector after digesting BsaI. So, you can perform digestion and ligation at the same time. You can obtain desired plasmids in a short time.

Figure 1 Golden Gate cloning strategy
2. Material & Method
We performed Golden Gate cloning with this part (vector) and mRFP (insert) and checked function. And we investigated the reaction rate changing mol ratio of vector to insert. The protocol is below.
1) PCR up insert with BsaI site
2) Golden Gate cloning
3) transformation
Mixture list in Golden Gate is below.
3. Result
Table 1 cfu/transformation

Table 2 Reaction ratio


Figure 2 Reaction ratio (N.D.: Not Detected)

Figure 3 Plate
In the traditional ligation, the best ratio is vecor: insert= 1: 3. However, according to this experiment, the best ratio was vector: insert= 1: 1in the Golden Gate. The vector ............................... vectorが切られたあと再びsfGFPとligateする可能性もあり,これがまた切られるためにBsaIが使用される。したがって,insertが多いとvectorとBsaIの衝突頻度が低下するため,liationが進みにくいと考えられる。
The maximum reaction rate was 68.9%. There existed few back ligations. Therefore, selecting colonies not shining green, you can pick the desired colonies easily.
CRISPRi
1.Introduction
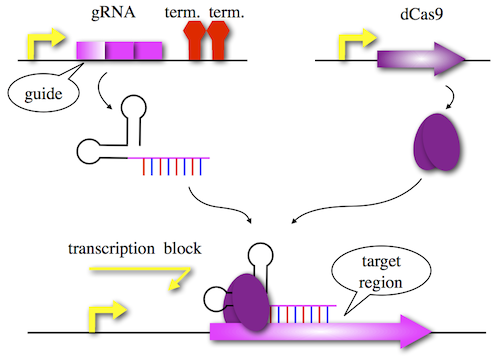
One of the immune system is CRISPR (clustered regularly interspaced short palindromic repeats). Cas9 protein and sgRNA (small guide RNA) combine specific sequence and cut it. Using a modified Cas9 lacking endonucleolytic activity, we can use CRISPR as repressor. This system is CRISPRi (CRISPR interference) as shown in Figure 1. Designing guide region of sgRNA and coexpress dCas9, you can knockdown target gene conditionally.
 "
"