Header
The nanoparticles were decided to be composed of gelatin, and a two step desolvation protocol was successfully used to obtain them. We were able to prove their presence by DLS method, giving an average size of nanoparticles of 200nm.
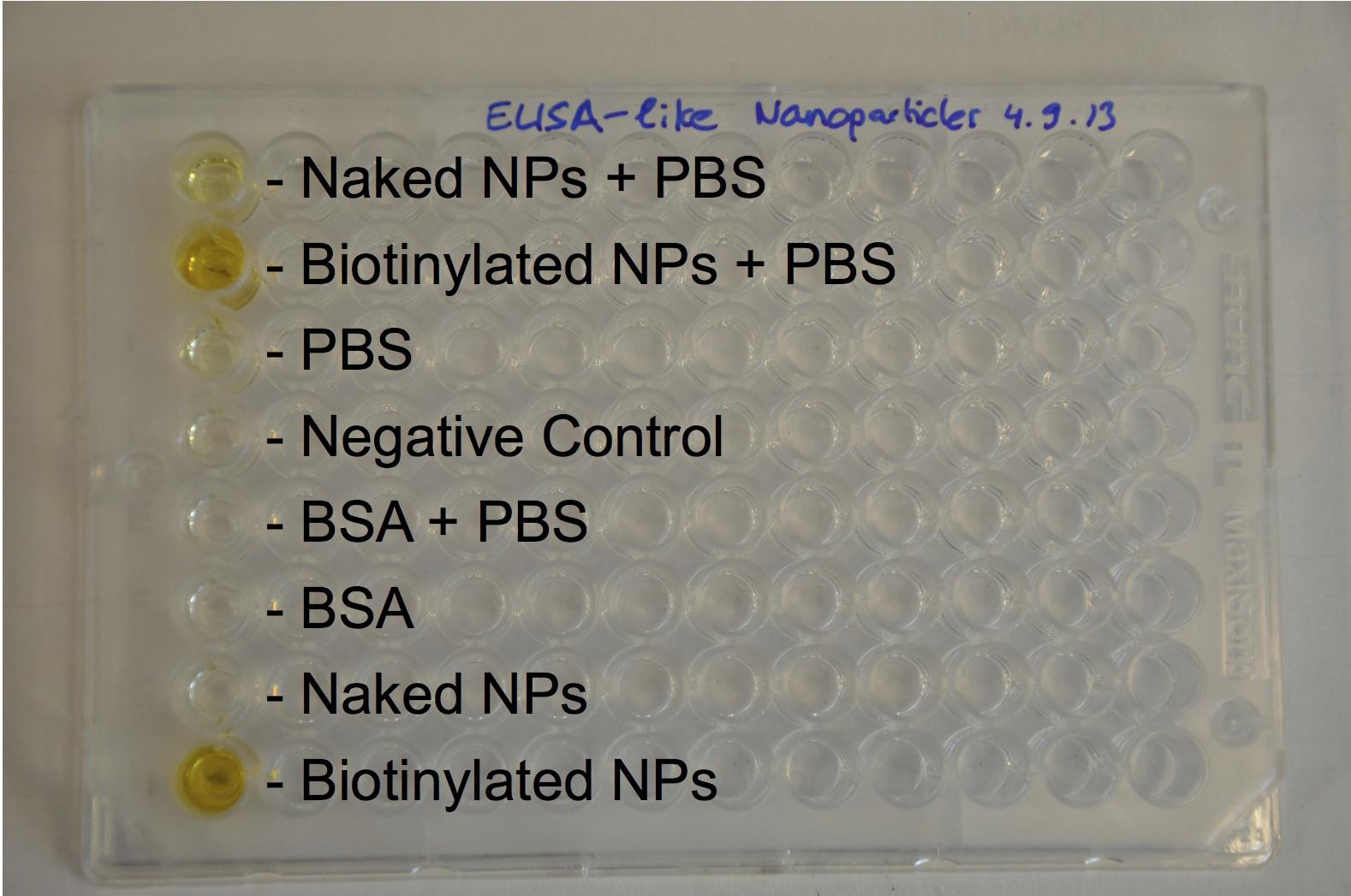
The biotinylation was chemically made with activated biotin and proved with an Elisa-like method.
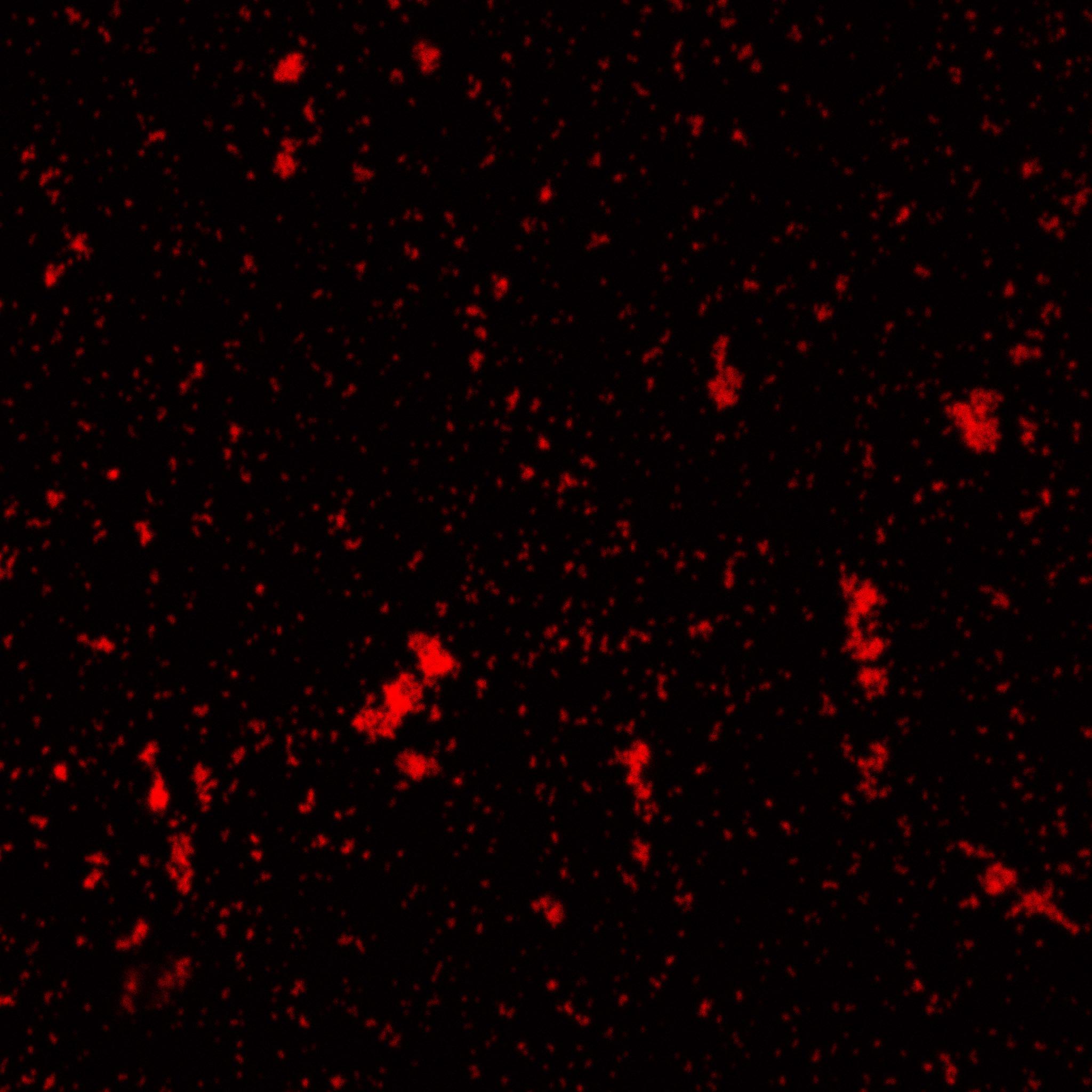
Biotinylated nanoparticles were dyed and observed under microscope.
Once proved that we could obtain nanoparticles, the loading was tried with two distinct fluorescent molecules : GFP, relatively small, and FITC-dextran, with a large MW due to sugars. FITC fluorescent images showed the incorporation of the molecule, while nanoparticles were not able to incorporate GFP, maybe due to its small MW or the acetone steps during the protocol.
Biotinylation was proved to be still possible after loading with FITC, what we did with an Elisa-like assay.
For sensing part, two pH-dependent and a constitutive promoter were inserted in front of a superfolded GFP sequence (Biobrick BBa_I746916 ). The inserts, consisting in the three different promoters, as well as the insert were isolated and amplified successfully by PCR. A gel was made to assess it and the sizes were the expected ones.
The Gibson assemblies went fine and the results matched 100% with the expected sequences.
Fluorescence measurements were not really relevant since a lot of cells died in acidic pH, supposed to activate the promoter. However, even if we were not able to prove that acidic pH triggers expression of GFP, fluorescence could be seen, thus the promoters are active. Constitutive promoter showed nice results, which allowed us to deliver it in the kit (see outreach -> kit).
For triggering expression of degradating enzymes, arabinose sensitive promoter was chosed. The enzymes to insert were MMP9, MMP2 and gelE, the three degrading gelatin. The part BBa_I746908 was the backbone consisting in GFP driven by pBad promoter. The enzymes to insert would be either put instead of GFP or in addition to GFP. All final plasmids would have a His-tag to purify it easily.
Sequences of the enzymes and the backbones were correctly PCR amplified. The Gibson were successfull for the MMP2 and gelE, but cells transformed with MMP9 didn't give any colony.
Sequencing results of the other plasmids showed a stop codon between enzymes and GFP, the reason why they didn't appear green.
However, the enzymes preceding GFP could still be expressed and an His-Tag purification was achieved since His-tag is placed before the stop codon. The different fractions were collected and analyzed on a SDS page :
Streptavidin export at the outer membrane was also needed to achieve our project in order to couple nanoparticles to bacteria. Three streptavidin were used to increase the success chances. The three were amplified and inserted into the Biobrick Bba_K523013 insted of YFP. So streptavidin was fused with INP supposed to export it at the outer membrane.
The sequencing results of the Gibson Assembly were fine and proved that streptavidin was well in frame with INP in the three cases.
The functionnal assays were less promising. An assay was performed with antibody anti streptavidin but showed no significant results. Biotin was then used because of its smaller size and showed .... on va le refaire demain/aujourd'hui.
A Western Blot was performed with a lot of unspecific noise, still showing some bands at the expected size of INP-streptavidin fusion (53KDa).
We also spent time on improving the part used as backbone for streptavidin export. Since the initial group didn't access a good microscop, we proved the location by performing an assay with antiYFP antibody, and it was proved to co-localize with the fluorescence obtained by excitaion of YFP, proving its export to the outer membrane.
In conclusion, we can say that except minor problems, cloning succedeed well. The Gibson assemblies globally worked out and we had no trouble growing the resulting transformed bacteria. The most delicate part was the characterization of our parts with functionnal assays. However, a lot of parts showed encouraging results but would maybe need to be studied in more detail. The naoparticles is the part that worked out well, nanoparticles could be synthetized and loaded. This project was ambitious and was almost achieved, and we are really proud of sharing our results with the iGEM community!
 "
"