Header
Sensing
Contents |
Overview
The Idea of this module was to transform the bacterium with a plasmid that would contain a promoter which senses a specific signal. Once this promoter senses the signal, it would initiate transcription of an enzyme which degrades the nanocapsule, thus releasing its contents. We decided to use pH as the specific trigger that activates the promoter. As a proof of principle, we inserted three different promoters into three plasmids in front of the bio brick BBa_I746916 which encodes superfolded GFP. Then we transformed cells with these plasmids and let them grow in media with different pHs in order to check the expression.
Experiments
We chose the following three pH sensitive promoters:
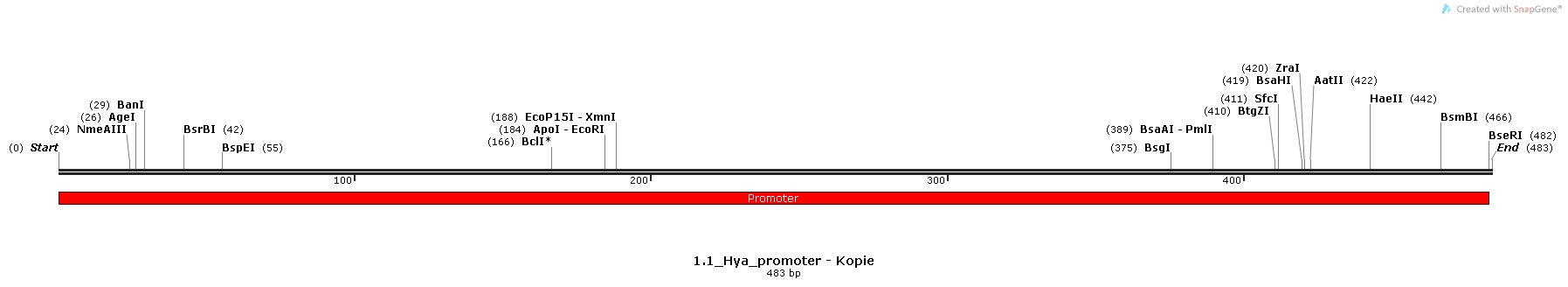
1.) Hya-promoter, isolated from the Escherichia Coli K-12 MG1655 strain
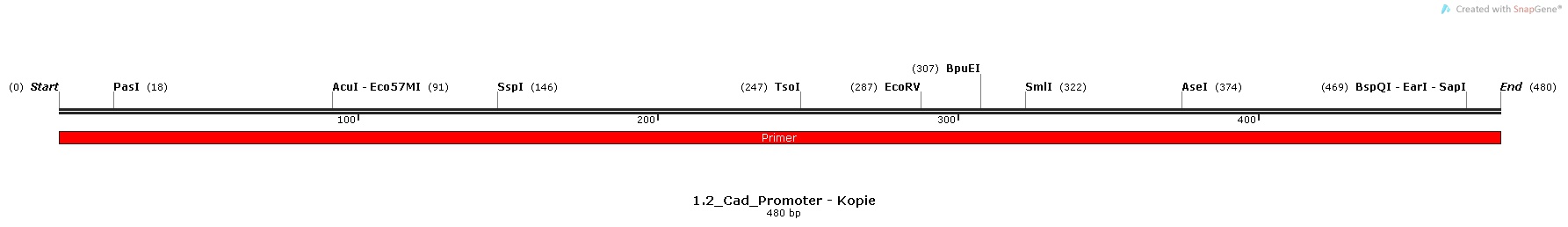
2.) Cad-promoter, isolated from the Escherichia Coli K-12 MG1655 strain
3.) BioBrick BBa_J23119, a constitutive promoter that was made by the 2006 Berkley team.
Promoter Sequences
Restriction Digest
We digested the Plasmid containing the biobrick BBa_I746908 which would serve us as backbone for our constructs.
PCRs and Gibson Assemblies
All the promoters were isolated by PCR and then assembled into the pSB1C3 Plasmid in front of the superfolded GFP.
Then each of the constructs was used to transform DH5-alpha competent cells, which were first platd on chloramphenicol containing LB agar plates for selection of positive transformants and then incubated into media with different pHs.
We used four media:
1.) LB-Chloramphenicol with a final pH of 5 (Adjusted with 10X MOPS+HCl)
2.) LB-Chloramphenicol with a final pH of 6 (Adjusted with 10X MOPS)
3.) LB-Chloramphenicol with a final pH of 7 (Adjusted with water)
4.) LB-Chloramphenicol with a final pH of 8.5 ( with 10X HEPES)
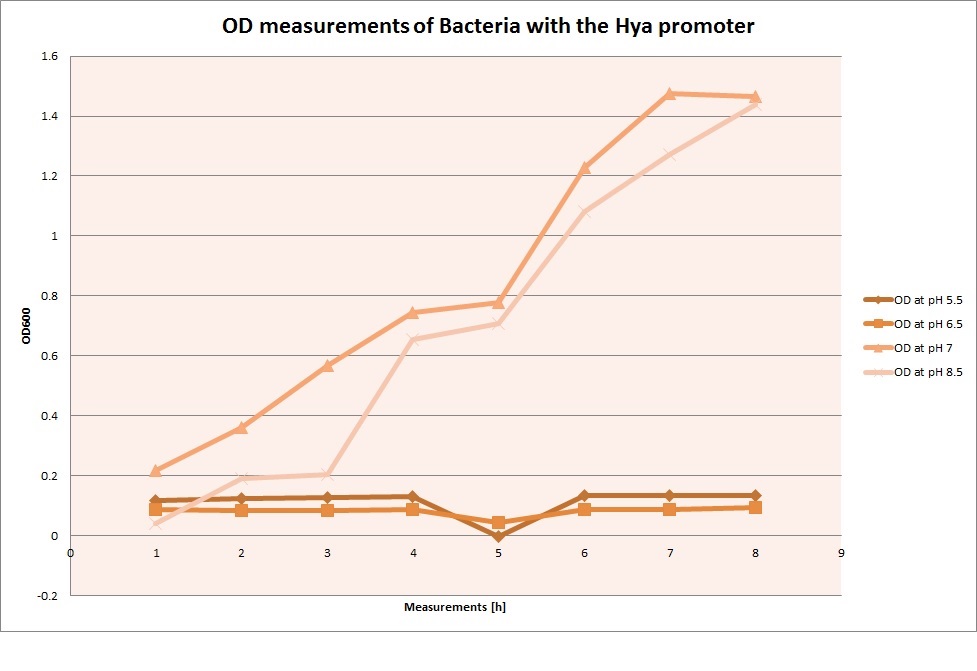
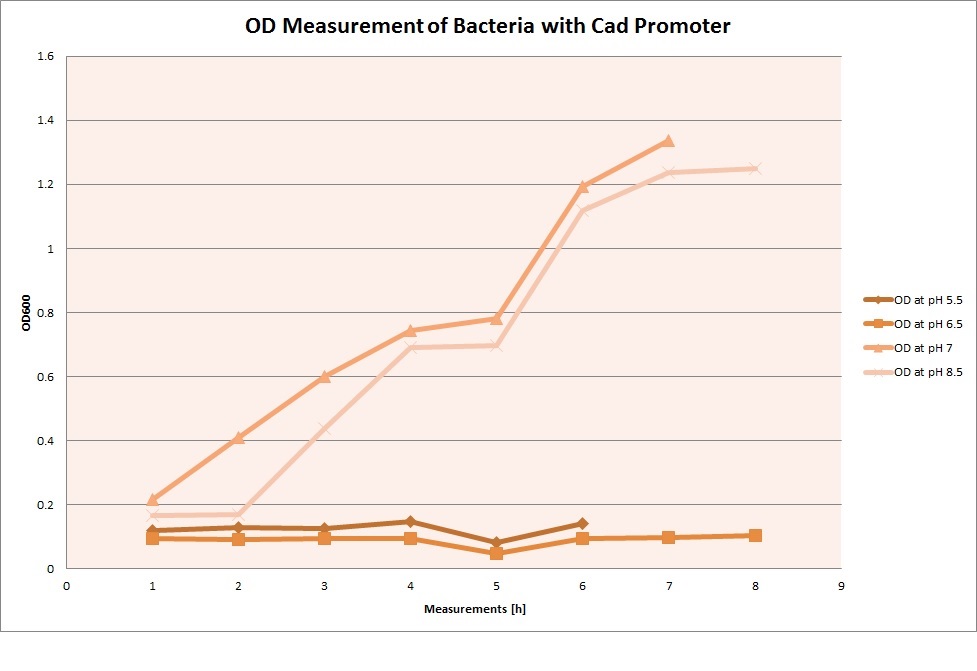
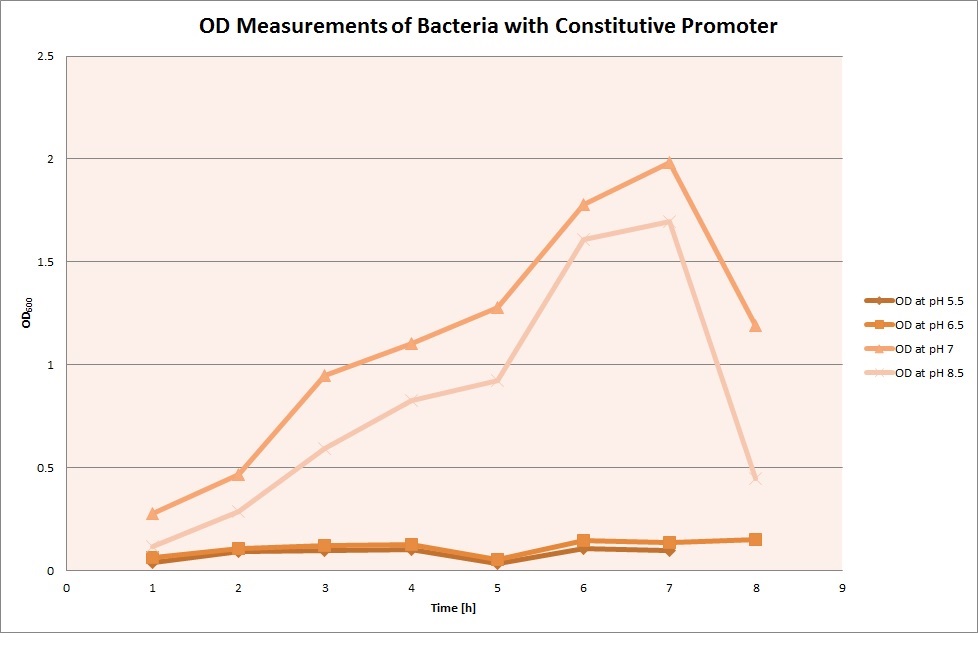
For each medium we measured the OD of the cells. This helped us to establish if a low expression of GFP was really due to the fact that the promoters did not work or simply that the cells were dying before they could initiate transcription and translation of the proten. It also would tell us if an increase of fluorescence was only due to an accumulation of bacteria or to the actual acumulation of GFP.
OD measurements
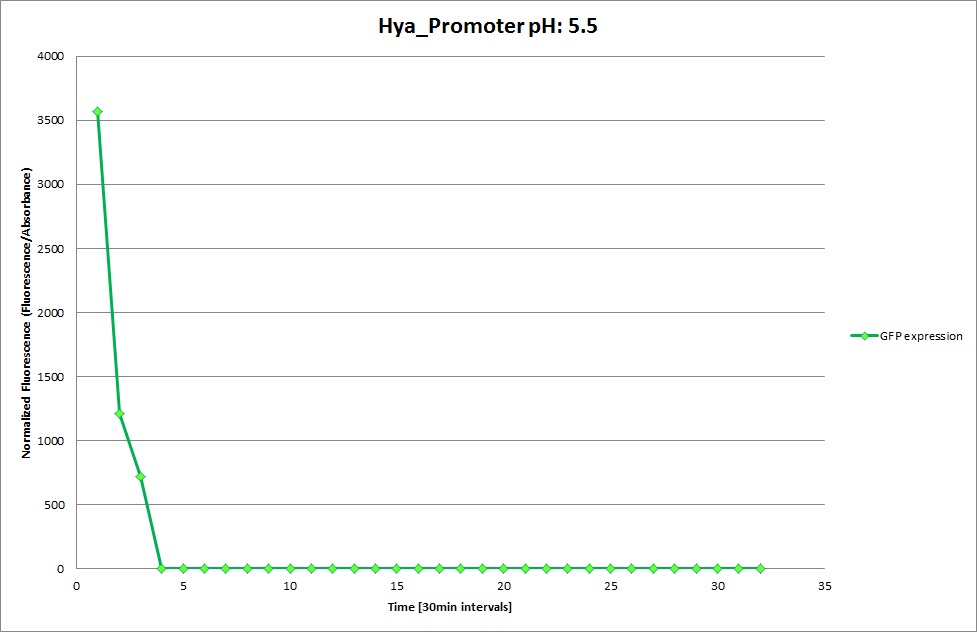
GFP expression was measured using a plate reader where we made three replicates for each promoter with each pH. Both the fluorescence as well as the 1:600 Absorbance was measured, in order to normalize the obtained fluorescence. Then, the average for each replicate was calculated and used to establish a curve depicting GFP expression within each medium.
GFP Measurements
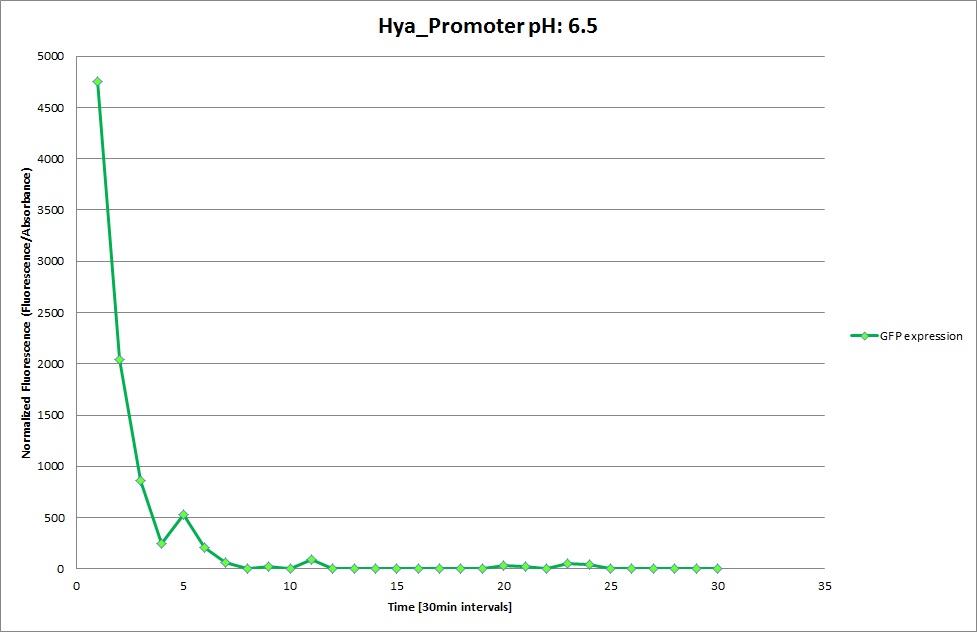
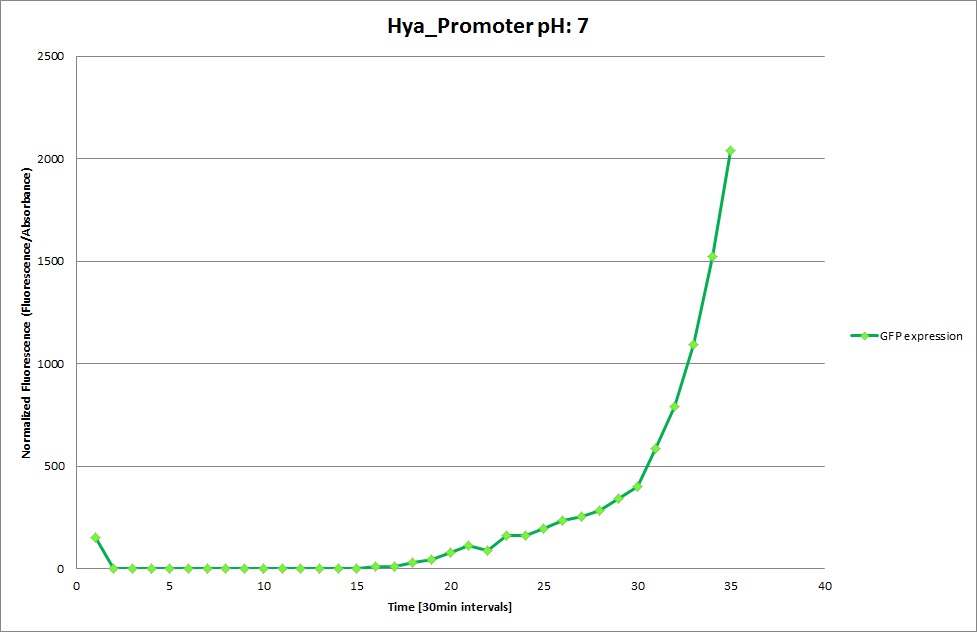
Hya Promoter
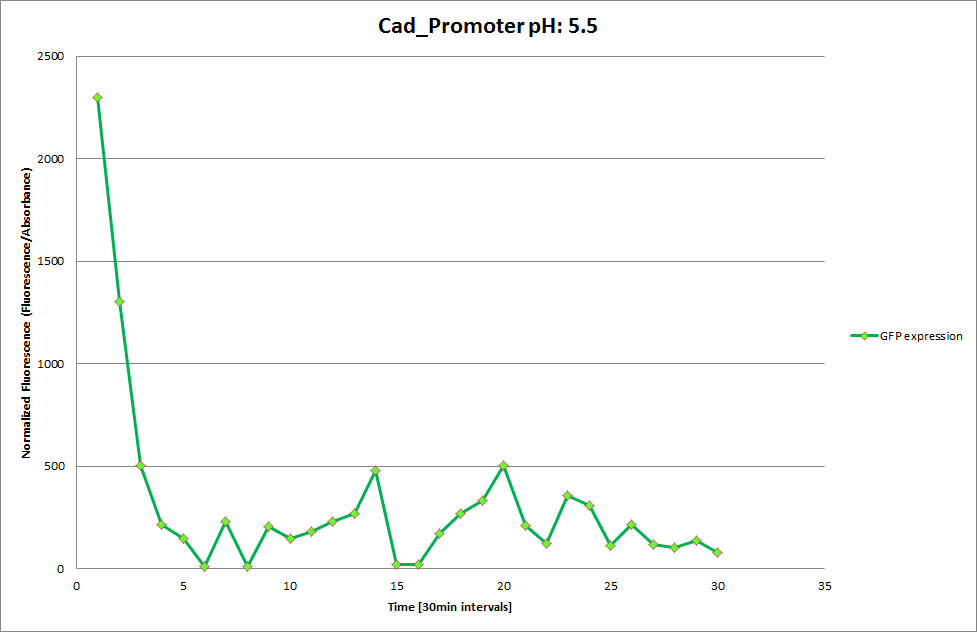
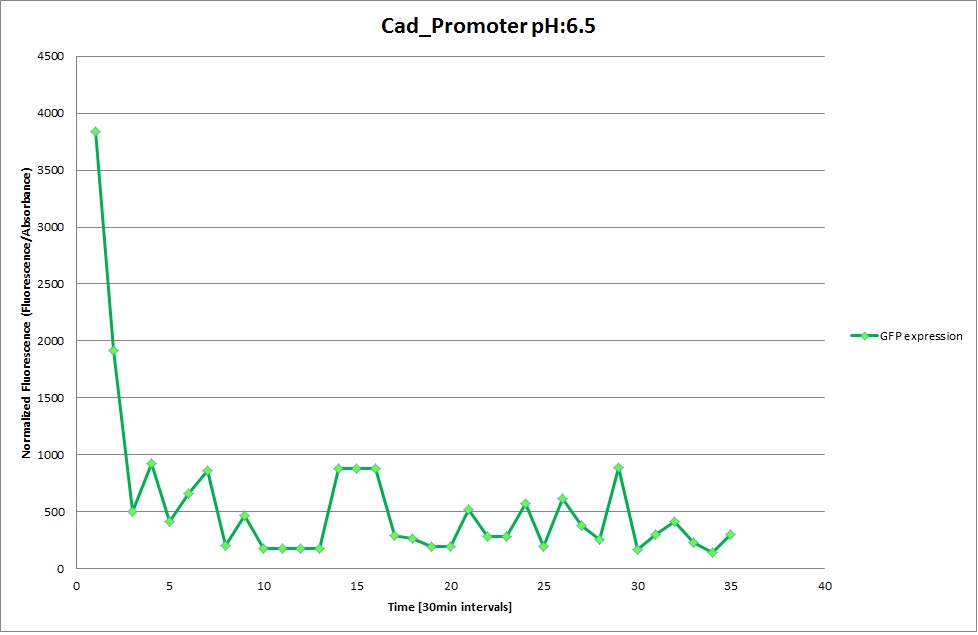
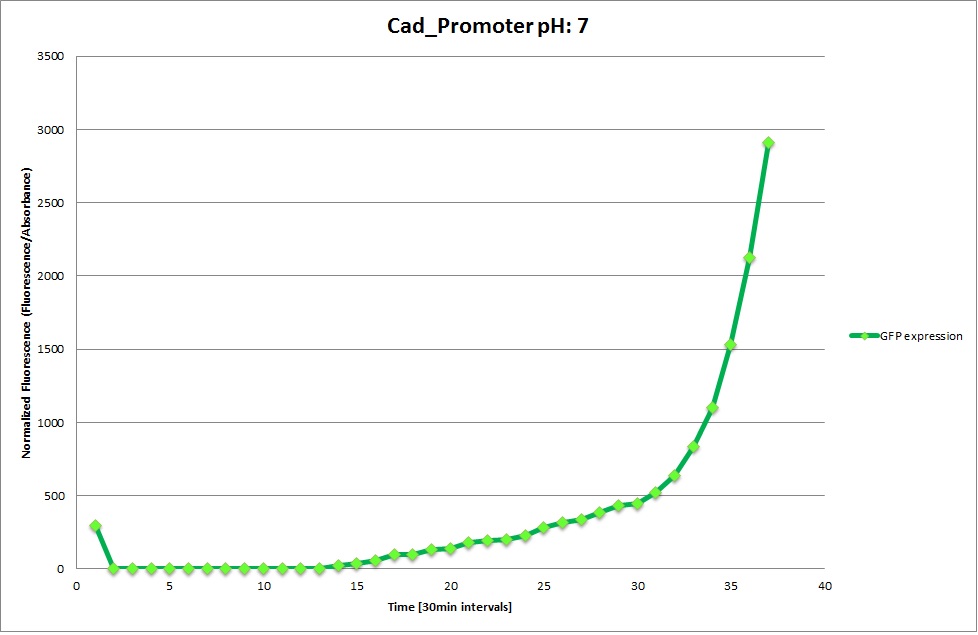
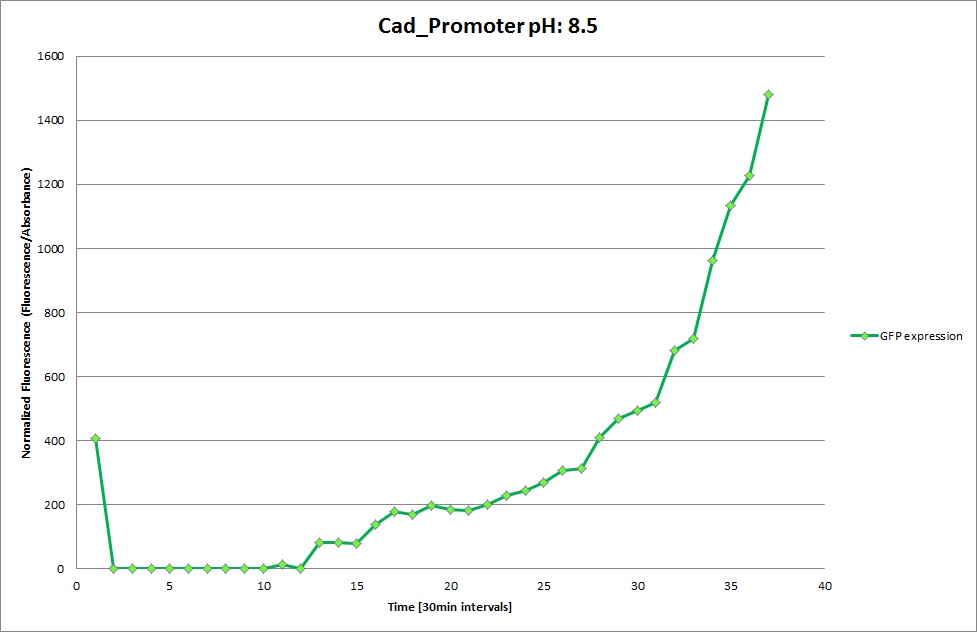
Cad Promoter
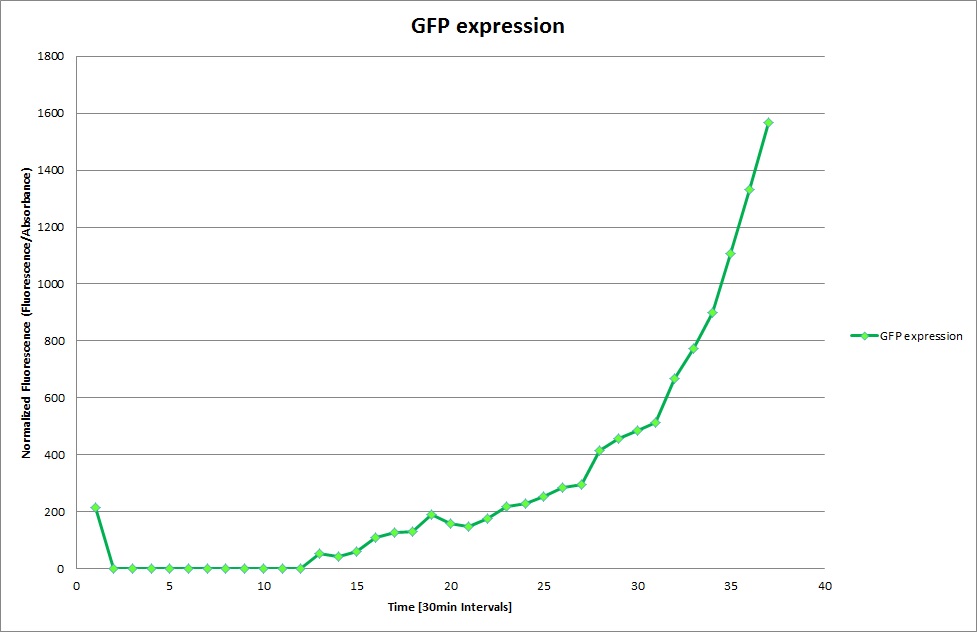
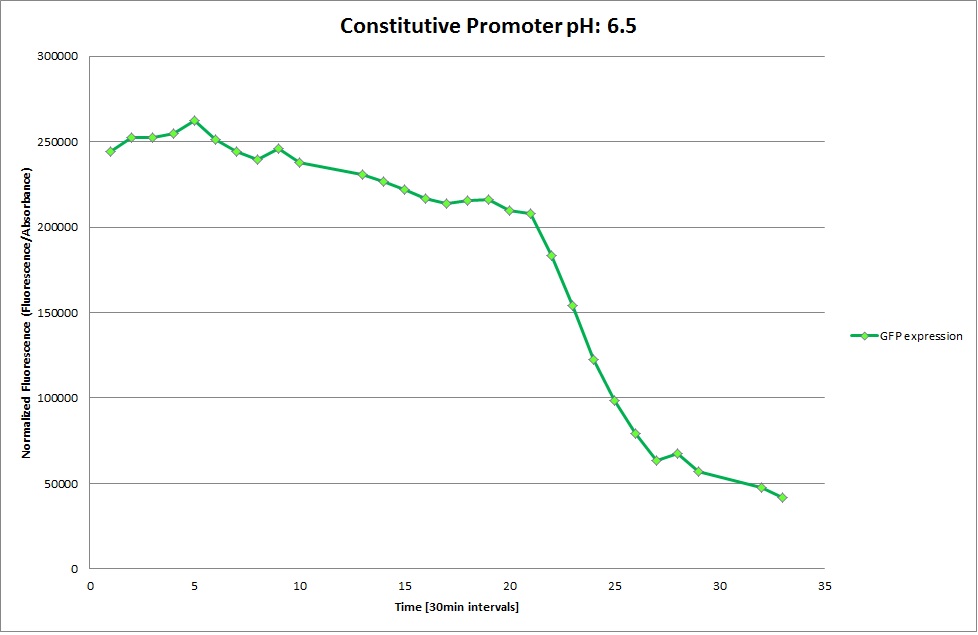
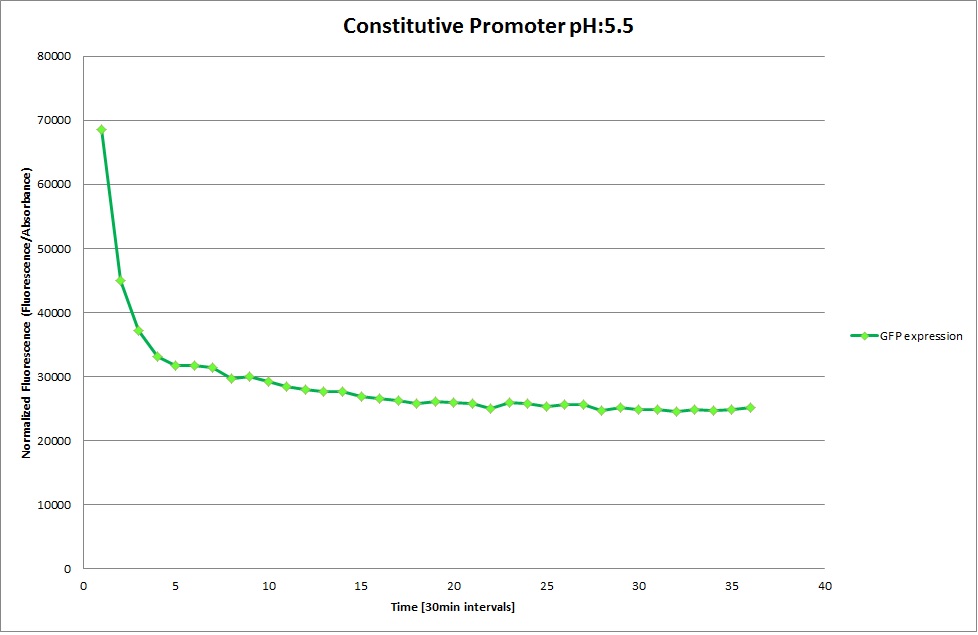
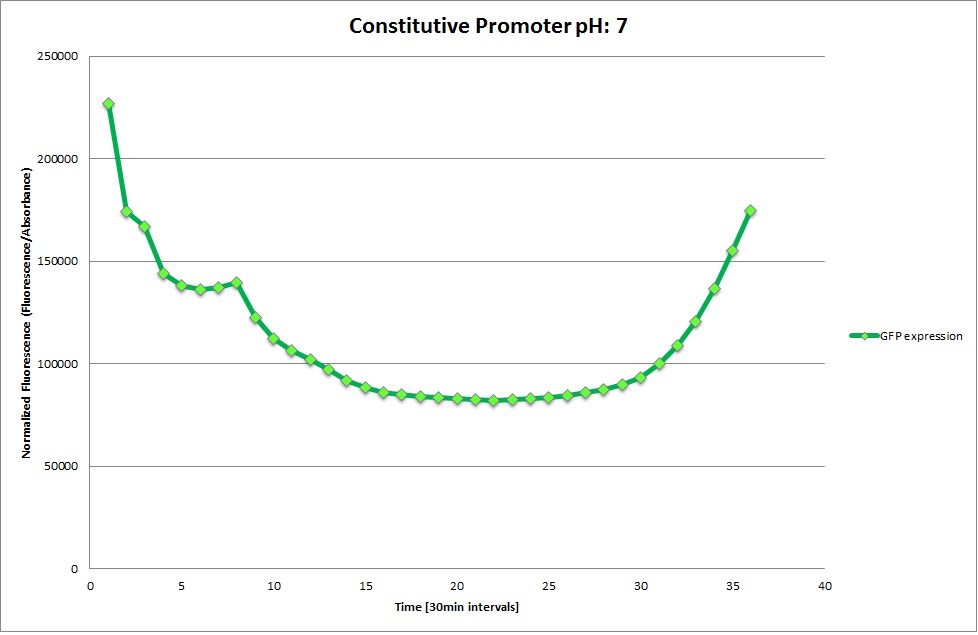
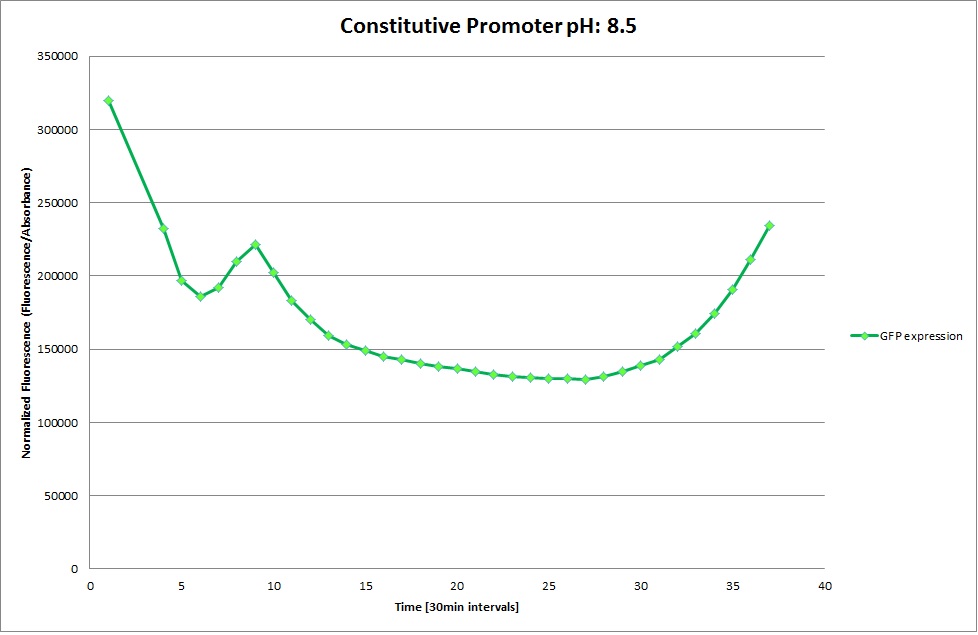
Constitutive Promoter
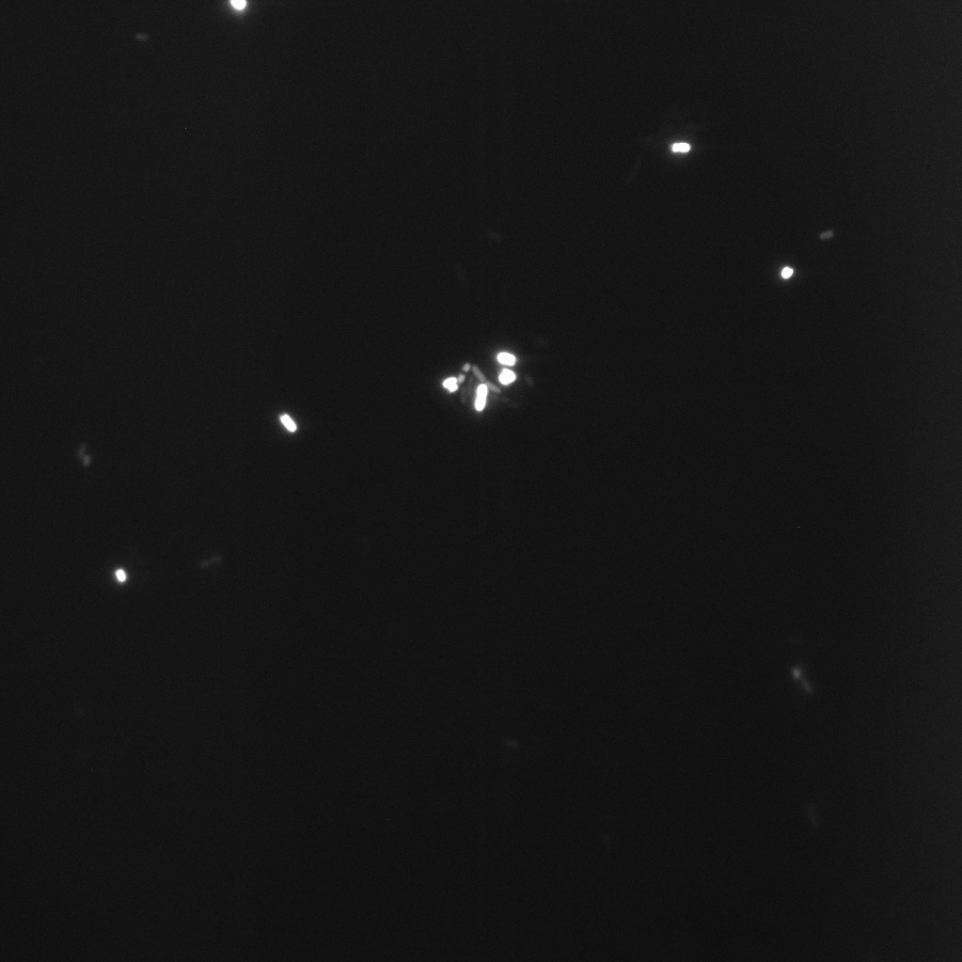
The Cells were also looked at under a microscope to qualitatively study their GFP expression.
Hya Promoter
Cad Promoter
Constitutive Promoter (Positive Control)
Expected outcome
The Promoters Hya and Cad were supposed to initiate transcription upon external acidification, which would then cause the bacteria to express superfolded GFP and turn bright green. The biobrick BBa_J23119, being a constitutive promoter, would turn the bacteria green independently of their medium. In respect to the project, the promoters would be inserted in front of a gelatinase so that the latter would be expressed upon arrival in an acidic medium. Here the bactrium would then secrete the gelatinase which would cause the degradation of the naonocapsule and the release of its contents.
Results
We successully managed to PCR amplify each of the three promoters as well as inserting them by Gibson Assembly into the pSB1C3 plasmid so that they would control the expression of superfolded GFP. The these assemblies were all successful, which was determined by sequencing the plasmid isolated from the positive trnasformants on the plate. This sequencing result showed a 100% match between the reference promoter sequence and the inserted sequence that the bacteria contained. Also, the bacteria containing the biobrick promoter had turned green on the plates as well as in liquid culture, which was just as expected.
Discussion
GFP expression under the hya and the cad promoters
From the measureents we made on the plate reader we were not able to conclude if the promoters are really induced at low pH as the cells died very quickly. This was also suported by the OD measurements we took. On those graphs one can see very clearly that the cells which are in the acidic media die almost immediately. Furhtermore there was some expression in both the neutral and the basic medium. This would indicate that the promoter is either a constitutive promoter or that it is inducible but leaky.
Comparing these graphs to the microscopy pictures one cannot say conclusively that the promoter is inducible by pH. Eventhough under the microscope there are only fluorescent bacteria at acidic pH the results from the plate reader clearly indicate that there are also fluorescent bacteria at neutral and high pH. One can see a trend though that for both the hya and the cad promoter the fluorescence is strongest at acidic pH and weakest at basic pH under the microscope. This trend cannot be seen in the PlateReader measurements, as the bacteria at low pH die before being able to express GFP.
Using the constitutive promoter as a positive control one can see that both hya and cad are weak promoters, as the fluorescence in the bacteria with the constitutive promoter is much stronger in acidic, neutral and basic media than the fluorescence in bacteria containing the hya or the cad promoter. Furthermore a comparison with the constitutive promoter again supports the hypothesis that the bacteria grow best at neutral pH, as there are much more bacteria at pH 7 and at pH 6.5 or 8.5.
Conclusion
Thus we can conclude that the hya promoter and the cad promoter work in the sense that they in fact induce expression of GFP. But from our data we were not able to conclusively say that they are inducible by low pH.
Nevertheless we were able to show that they are weak promoters that can be used to express a protein that should be present in low concentrations.
With respect to our project
In order to be sure that hya and cad are induced only at low pH we would need to do further testing. As soon as their inducibility would be confirmed we would insert them by gibson assemly in front of either GelE gelatinase or MMP2 gelatinase. Upon arrival in a medium with pH bellow 7 the enzyme would be synthetised and released.
References:
Journal of Bacteriology
Response of hya Expression to External pH in Escherichia coli
Paul W. King and Alan E. Przybyla
J.Bacteriol. 1990
Journal of Bacteriology
Mutational analysis and charecterization of the Escherichia coli hya operin, which encodes [NiFe] hydrogenase1
N K Menon, J Robbins, J C Wendt, K T Shanmuagam and A E Przybyla
J. Bacteriol 1991
Journal of Bacteriology
Nucleotode Sequence of the Escherichiea coli cad Operon: a system for neutralization of Low Extracellular pH
Shi-Yuan meng and George N. Bennett
J. Bacteriol 1992
Journal of Bacteriology
Regulation of the Escherichia coli cad Operon: Location of a Site Required for Acid Induction
Shi-Yuan meng and George N. Bennett
J. Bacteriol 1992
Effecting
Overview:
The goal of the effecting part of the project was to build a plasmid containing an arabinose promoter driving a tagged enzyme able to cleave gelatinase, the fabric used for our nano particles. We chose three different enzymes that would be inserted in part BBa_I746908, a pBAD promoter driving superfolder GFP, either between the promoter and GFP tag or instead of the GFP tag since it sometimes causes folding troubles. All of the constructs were planned to have a His tag as well, just in case something went wrong with the GFP tag, so we could extract and purify our expressed protein anyway.
Experiments:
The three different enzymes that we chose to use were gelatinase E (gelE) from Gram positive bacterium E. faecalis, metalloprotease 2 (MMP2) from H. sapiens and metalloprotease 9 (MMP9) from M. musculus. The genomic DNA of E. faecalis was ordered and a PCR was performed on it using Gibson primers that also added a His tag in front of the protein sequence. For MMP2 and MMP9, plasmids containing the coding sequence (CDS) of the proteins were ordered from plasmid.med.harvard.edu and the CDS were extracted in the same way as for gelatinase. In the meantime, PCRs were performed on the iGEM part BBa_I746908 to clone the backbone, add the overlap for the Gibson and, when needed, remove the GFP from the backbone (since we wanted to have a construct for each protein where GFP was replaced directly by the CDS). Competent cells were then transformed with the successful constructs, grown at 37C and later on activated, by adding 50ul of 20% arabinose solution to 5ml of inoculated culture, grown overnight, for 2h. A Western blot was then performed on the lysate and supernatural of the cells to check for the presence of the protein either out (if secreted as desired) or in (not secreted but at least expressed) the cells. A His-tag purification followed by SDS-PAGE were also executed.
Expected outcome:
We would expect the cells to be green and express/secrete a protein-GFP complex upon arabinose induction if everything works perfectly. The secreted protein, if correctly folded, could then break open the nano particles attached to the bacteria's surface, thus releasing the contained drug. However, we did not proceed to transform our bacteria with the coupling plasmid since the coupling section was stopped due to the lack of an antibody needed to prove that the coupling plasmid was complete.
Results:
The PCR reactions to amplify and extract the proteins CDSs and plasmid backbone were successful. We then proceeded to a Gibson assembly of the constructs that were meant to keep the GFP, for all three proteins. The Gibsons for the pBAD/araC-gelE-GFP and pBAD/araC-MMP2-GFP constructs worked, but not the one for the MMP9. We then transformed bacteria, isolated the plasmids and sent them for sequencing with the iGEM primers for the backbone and also with our own Gibson primers. This showed that the Gibson had been a success, but that a STOP codon had appeared in front of the GFP sequence, just after the linker. Indeed, the transformed bacteria did not appear green when induced with arabinose. Since a His tag had been added to the CDS of each protein using the Gibson primers, we did a Western blot with anti-his tag antibodies on the induced cell media and lysates. The result came out negative. We then proceeded to a his-tag purification of the cell lysates (Ni-NTA spin kit).
 "
"