Header
Sensing
Overview
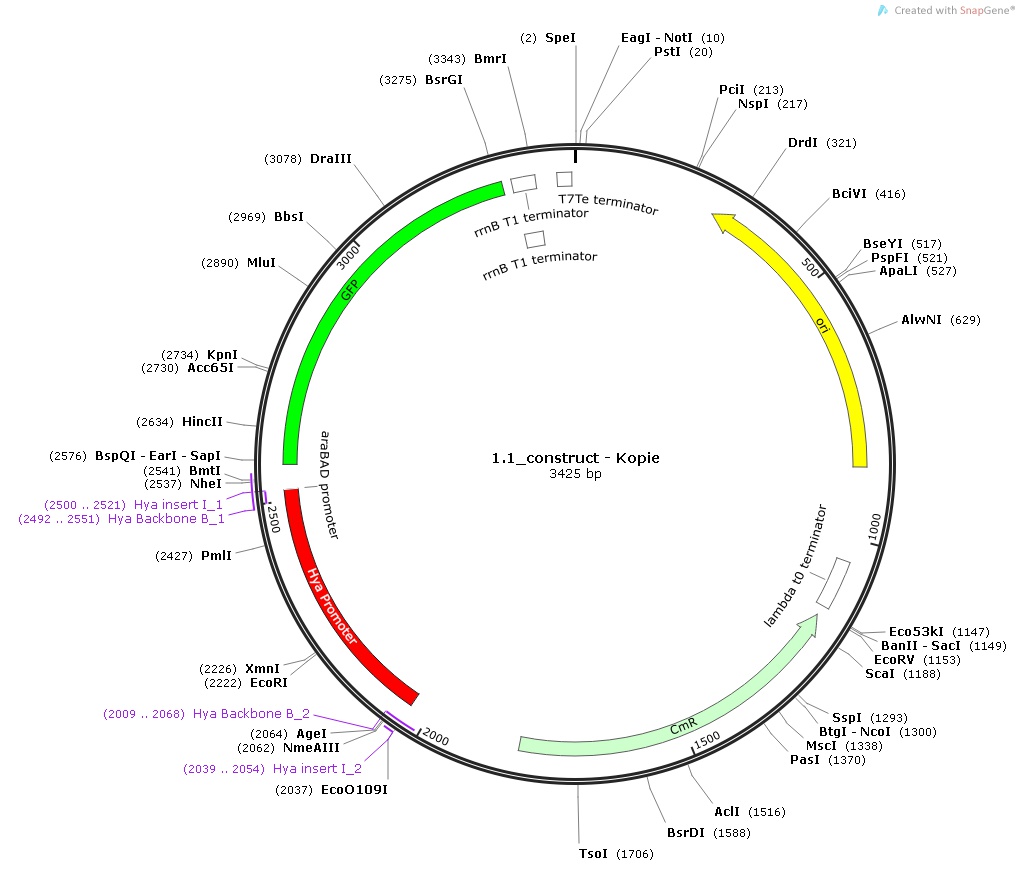
The Idea of this module was to transform the bacterium with a plasmid that would contain a promoter which senses a specific signal. Once this promoter senses the signal, it would initiate transcribtion of an enzyme which degrades the nanocapsule, thus releasing its contents. We decided to use pH as the specific trigger that activates the promoter. As a proof of principle, we inserted three different promoters into three plasmid in front of the bio brick BBa_I746916 which encodes superfolded GFP. Then we transformed cells with these plasmids and let them grow in media with different pHs in order to check the expression.
Experiments
We chose the following three pH sensitive promoters:
1.) Hya-promoter, isolated from the Escherichia Coli K-12 MG1655 strain
2.) Cad-promoter, isolated from the Escherichia Coli K-12 MG1655 strain
3.) BioBrick BBa_J23119, a constitutive promoter that was make by the 2006 Berkley team.
All the promoters were isolated by PCR and then assembled into the pSB1C3 Plasmid in front of the superfolded GFP. Then each of the constructs was used to transform DH5-alpha competent cells which were first plated and then incubated into media with different pHs.
We used four media:
1.) LB-Chloramphenicol with 10X MOPS+HCl, with a final pH of 5
2.) LB-Chloramphenicol with 10X MOPS, with a final pH of 6
3.) LB-Chloramphenicol without any buffer, with a final pH of 7
4.) LB-Chloramphenicol with 10X HEPES, with a final pH of 8.5
GFP expression was measured using a plate reader where we made three replicates of each promoter with each pH. Then the average for each replicate was calculated and used to establish a curve depicting GFP expression within each medium. Expected outcome:
The Promoters Hya and Cad were supposed to initiate transcription upon external acidification, which would then cause the bacteria to express superfolded GFP and turn bright green. The biobrick from 2006, being a constitutive promoter, would turn the bacteria green independent of their medium.
Results:
We successully managed to isolate and amplify each of the three promoters as well as inserting them into the pSB1C3 plasmid so that they would controll the expression of superfolded GFP. The transformation that followed was also successful which was determined by sequencing the plasmid isolated from the colonies on the plate. This sequencing result showed a 100% match between the original promoter sequence and the inserted sequence that the bacteria contained. Also, the bacteria containing the biobrick pH-promoter had turned green on the plates as well as in liquid culture, which was just as expected.
 "
"