Team:Heidelberg/Tyrocidine week20 biobrickmod
From 2013.igem.org
Detection of short synthetic NRPs
03.09.2013 Pre-test
Pre-test: solubilize the synthetic dipeptide in Methanol; result: very bad solubility => not used for MS comparisons
03.09. - 04.09.2013 Preparation of samples 1-10 for mass spectrometry at neonate screening
Inocculation of BAP1 (Dipeptide I) and BAP1 (Tripeptide I) [Both tested on NRPS content with SDS-PAGE] in LB media. Washing and transfer of cells to M9 minimal media (evening 01.09.2013) and IPTG induction (noon 02.09.2013) of 50 ml (each) culture.
Taking first 15 ml samples after 2.5 hours. Centrifugation (3750 rpm, 4°C, 45 min) and separation of pellet and supernatant (2x ~7,5ml). Freezing with nitrogen and Lyophilization for 12 hours at ~-45°C and ~0,1 mbar.
Second sample (taken 12 hours after induction) and third one (21 hours after induction) have been treated as the first one.
Inocculation of native BAP1 cells in LB medium, washing in M9 and growing of cultures in M9 as negative reference for mass spec. Same treatment as positive samples.
| Sample number | Content | MS- Ornithine content [µmol/L] |
|---|---|---|
| 1 | Di A in BAP1 in M9+Cm Pellet 21 hrs after induction | 6,05 |
| 2 | Tri I in BAP1 in M9+Cm Pellet 12 hrs after induction | 29,81 |
| 3 | Tri I in BAP1 in M9+Cm Pellet 21 hrs after induction | 40,24 |
| 4 | Tri I in BAP1 in M9+Cm Pellet 2.5 hrs hrs after induction | 27,72 |
| 5 | Di A in BAP1 in M9+Cm Pellet 2.5 hrs after induction | 6,49 |
| 6 | Di A in BAP1 in M9+Cm Pellet 12 hrs after induction | 4,77 |
| 7 | Di A in BAP1 M9+cm Medium 1 2.5 hrs after induction | 4,78 |
| 8 | Tri I in BAP1 M9+Cm Medium 1 2.5 hrs after induction | 166,07 |
| 9 | Di A in BAP1 M9+Cm Medium 2 2.5 hrs after induction | 7,09 |
| 10 | Tri 1 in BAP1 M9+Cm Medium 2 2.5 hrs after induction | 173,36 |
Samples 1-6 contained different pellets, which have been resuspended in 500 µL PBS Buffer (1X) and treated with 3x20[s] ultra-sonification. Finally the samples have been centrifuged and supernatant decanted to 1.5 ml tubes for mass spectrometry at neonate screening.
Samples 7-10 contained lyophilized Medium and have been resuspended in 500 µL methanol for MS screening. As samples 1-6, supernatant was separated from solid contents and decanted to 1.5ml tubes. 100 µL were handed for MS at neonate screening. The rest has been frozen for hydrolysis.
05.09.-06.09.2013 Preparation of samples 11-21 for mass spectrometry at neonate screening
Samples treated as samples 1-10.
| Sample number | Content | MS result (Ornithine) |
|---|---|---|
| 11 | Di A in BAP1 M9+Cm after 12 hrs after induction Medium 1 | 15,5 |
| 12 | Di A in BAP1 M9+Cm after 21 hrs after induction Medium 1 | 10,2 |
| 13 | Di A in BAP1 M9+Cm after 21 hrs after induction Medium 2 | 10,7 |
| 14 | Di A in BAP1 M9+Cm after 21 hrs after induction Medium 3 | 7,8 |
| 15 | TriI in BAP1 M9+Cm after 12 hrs after induction Medium 1 | 1138,0 |
| 16 | TriI in BAP1 M9+Cm after 21 hrs after induction Medium 1 | 879,1 |
| 17 | TriI in BAP1 M9+Cm after 21 hrs after induction Medium 2 | 1343,4 |
| 18 | TriI in BAP1 M9+Cm after 21 hrs after induction Medium 3 | 1324,4 |
| 19 | BAP1 native M9 after 12 hrs after induction Medium 1 | 73,8 |
| 20 | BAP1 native M9 after 12 hrs after induction Medium 2 | 18,4 |
| 21 | BAP1 native M9 after 12 hrs after induction Pellet | 8,2 |
Samples with "H" are hydrolysates (6 M HCL at 105° over night). Those were hydrolysed over night, evaporated and neutralized for MS. Resolved supernatant for "media" samples has been analysed separately for the methanol supernatant and solid phase (marked with "sol").
| Sample number | Content | MS result |
|---|---|---|
| H 1 | Di A in BAP1 in M9+Cm Pellet 21 hrs after induction | not needed |
| H 2 | Tri I in BAP1 in M9+Cm Pellet 12 hrs after induction | not needed |
| H 3 | Tri I in BAP1 in M9+Cm Pellet 21 hrs after induction | not needed |
| H 4 | Tri I in BAP1 in M9+Cm Pellet 2.5 hrs after induction | not needed |
| H 5 | Di A in BAP1 in M9+Cm Pellet 2.5 hrs after induction | not needed |
| H 6 | Di A in BAP1 in M9+Cm Pellet 12 hrs after induction | not needed |
| H 7 | Di A in BAP1 M9+cm Medium 1 2.5 hrs after induction | not needed |
| H 8 | Tri I in BAP1 M9+Cm Medium 1 2.5 hrs after induction | not needed |
| H 9 | Di A in BAP1 M9+Cm Medium 2 2.5 hrs after induction | not needed |
| H 10 | Tri 1 in BAP1 M9+Cm Medium 2 2.5 hrs after induction | not needed |
| H 11 | Di A in BAP1 M9+Cm 12 hrs after induction Medium 1 | not needed |
| H 11 sol | Di A in BAP1 M9+Cm 12 hrs after induction Medium 1 | not needed |
| H 12 | Di A in BAP1 M9+Cm 21 hrs after induction Medium 1 | not needed |
| H 12 sol | Di A in BAP1 M9+Cm 21 hrs after induction Medium 1 | not needed |
| H 13 | Di A in BAP1 M9+Cm 21 hrs after induction Medium 2 | not needed |
| H 13 sol | Di A in BAP1 M9+Cm 21 hrs after induction Medium 2 | not needed |
| H 14 | Di A in BAP1 M9+Cm 21 hrs after induction Medium 3 | not needed |
| H 14 sol | Di A in BAP1 M9+Cm 21 hrs after induction Medium 3 | not needed |
| H 15 | TriI in BAP1 M9+Cm 12 hrs after induction Medium 1 | not needed |
| H 15 sol | TriI in BAP1 M9+Cm 12 hrs after induction Medium 1 | not needed |
| H 16 | TriI in BAP1 M9+Cm 21 hrs after induction Medium 1 | not needed |
| H 16 sol | TriI in BAP1 M9+Cm 21 hrs after induction Medium 1 | not needed |
| H 17 | TriI in BAP1 M9+Cm 21 hrs after induction Medium 2 | not needed |
| H 17 sol | TriI in BAP1 M9+Cm 21 hrs after induction Medium 2 | not needed |
| H 18 | TriI in BAP1 M9+Cm 21 hrs after induction Medium 3 | 273,3 |
| H 18 sol | TriI in BAP1 M9+Cm 21 hrs after induction Medium 3 | not needed |
| H 19 | BAP1 native M9 12 hrs after induction Medium 1 | not needed |
| H 19 sol | BAP1 native M9 12 hrs after induction Medium 1 | not needed |
| H 20 | BAP1 native M9 12 hrs after induction Medium 2 | not needed |
| H 21 | BAP1 native M9 12 hrs after induction Pellet | not needed |
09.09.-11.09.2013 further preparation for MS at chemistry
Preparation of cultures containing di- and tripeptide for MS (inocculation of LB, washing in M9, transfer to M9 and IPTG induction). IPTG induction at 12 p.m. 10.09., samples taken after 8.5 and 24 and 48 hours. Procedure as mentioned above (lysis, lophylisation etc.), but took pellet from 10 ml and lyophilized 7.5 ml supernatant again.
Results of hydrolysis (05.09.-06.09): A Maillard reaction occurred! Everything brown and many different products (as bonds between amino acids and sugars) arised during cooking in HCL (this protocol seems to be only used for pure peptide solutions). Therefore we have to search for other enzymatic reactions or other strategies to assess ornithine concentrations. Tried to rescue samples by an alkaline reaction with 5 M NaOH, but samples got only slightly lighter.
| Sample number | Content | MS result |
|---|---|---|
| 22 | DiA in BAPI Pellet in M9+Cm+IPTG 8.5 hrs after induction 10.09.2013 | 13,0 |
| 23 | TriI in BAPI Pellet in M9+Cm+IPTG 8.5 hrs after induction 10.09.2013 | 33,5 |
| 24 | Di A in BAPI Pellet1 in M9+Cm+IPTG 24 hrs after induction 11.09.2013 | 19,4 |
| 25 | TriI in BAPI Pellet1 in M9+Cm+IPTG 24 hrs after induction 11.09.2013 | 88,0 |
| 26 | Di A in BAPI Pellet2 in M9+Cm+IPTG 24 hrs after induction 11.09.2013 | 11,2 |
| 27 | TriI in BAPI Pellet2 in M9+Cm+IPTG 24 hrs after induction 11.09.2013 | 119,4 |
| 28 | DiA in BAPI Medium in M9+Cm+IPTG 8.5 hrs after induction 10.09.2013 | 7,8 |
| 29 | TriI in BAPI Medium in M9+Cm+IPTG 8.5 hrs after induction 10.09.2013 | 582,1 |
| 30 | Di A in BAPI Medium1 in M9+Cm+IPTG 24 hrs after induction 11.09.2013 | 13,9 |
| 31 | Di A in BAPI Medium2 in M9+Cm+IPTG 24 hrs after induction 11.09.2013 | 6,3 |
| 32 | TriI in BAPI Medium1 in M9+Cm+IPTG 24 hrs after induction 11.09.2013 | 520,2 |
| 33 | TriI in BAPI Medium2 in M9+Cm+IPTG 24 hrs after induction 11.09.2013 | 504,5 |
| 34 | Di A in BAPI Pellet in M9+Cm+IPTG 48 hrs after induction 12.09.2013 | 263,3 |
| 35 | Di A in BAPI Medium in M9+Cm+IPTG 48 hrs after induction 12.09.2013 | 498,2 |
| 36 | TriI in BAPI Pellet in M9+Cm+IPTG 48 hrs after induction 12.09.2013 | 373,7 |
| 37 | TriI in BAPI Medium in M9+Cm+IPTG 48 hrs after induction 12.09.2013 | 356,6 |
12.09.further preparation and delievery to MS at chemistry
Resuspension of lyophilized samples in 500 microliters methanol. Delievery of samples 25 and 26 to MS. Other sam==RFC10 Standardization of Tyc Modules==
Sequencing
| Primer | OK? | Comments | File |
|---|---|---|---|
| Tyc AdCom | |||
| VR_rv | - | ||
| VF2_fw | - | ||
| Tyc B1 | |||
| VR_rv | silent mutations: 4595 GCC (Ala) to GCG (Ala); 4598 CTT (Leu) to CTG (Leu) | ||
| VF2_fw | |||
| Tyc C5 | |||
| VR_rv | backbone was ok, wrong insert; since the insert was amplified with the same protocol like TycB1 we think that the primers were mixed up | ||
| VF2_fw | backbone was ok, wrong insert; since the insert was amplified with the same protocol like TycB1 we think that the primers were mixed up | ||
| Tyc C6 | |||
| VR_rv | |||
| VF2_fw | silent mutations: 4595 GTA (Val) to GTG (Val) | ||
Tyrocidine-Indigoidine-fusion - standardization
So far, the Tyrocidine-Indigoidine-fusion constructs had two illegal cutting sites of RFC-10 restriction enzymes. Hence, we tried a CPEC-approach for which we reamplified a fragment from the vector with primers that introduced a mutation at the desired position.
Amplification
| what | µl |
|---|---|
| pPW05 (dil.) | 1 |
| RB68 | 2 |
| RB69 | 2 |
| Phusion Flash 2x Master Mix | 10 |
| ddH20 | 5 |
| Cycles | temperature [°C] | Time [min:s] |
|---|---|---|
| 1 | 98 | 0:05 |
| 35 | 98 | 0:05 |
| 59 | 0:10 | |
| 72 | 3:00 | |
| 1 | 72 | 10:00 |
| 1 | 10 | inf |
Results
CPEC
After gel-extraction of the large fragment, the following procedure was followed:
- 22µl H2O, 20µl of the large fragment, 2µl of the short fragment, 1µl DpnI and 5µl 10x CutSmart buffer were incubated for 2 hours at 37°C
- Mixture was purified with isoprop, washed with ethanol and eluted in 10µl H2O
- 10µl Phuson Flash Master Mix was added and the followin protocol was run:
| Cycles | temperature [°C] | Time [min:s] |
|---|---|---|
| 1 | 98 | 0:10 |
| 5 | 98 | 0:01 |
| 53 | 0:05 | |
| 72 | 3:00 | |
| 1 | 72 | 10:00 |
| 1 | 10 | inf |
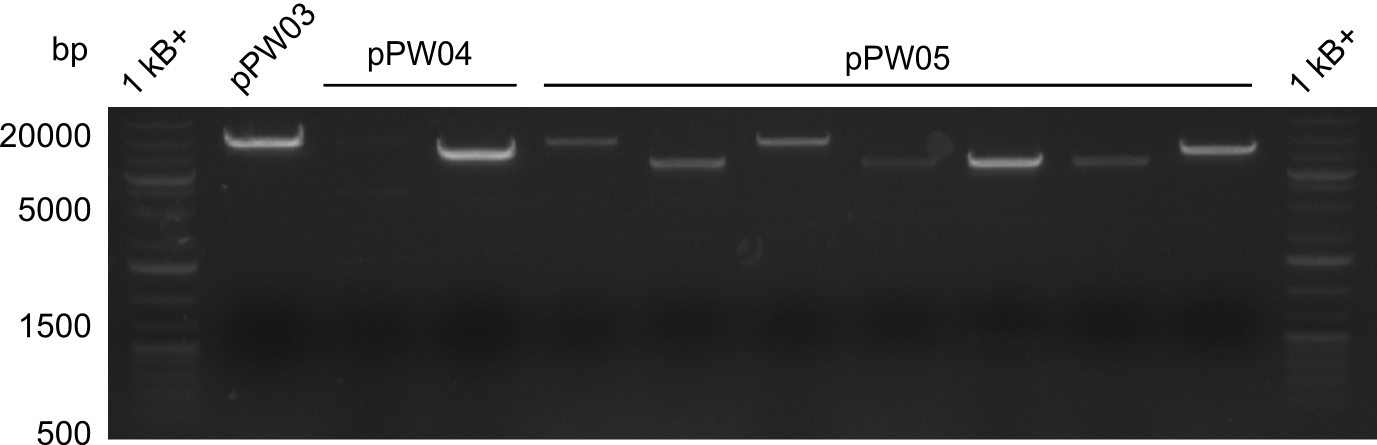
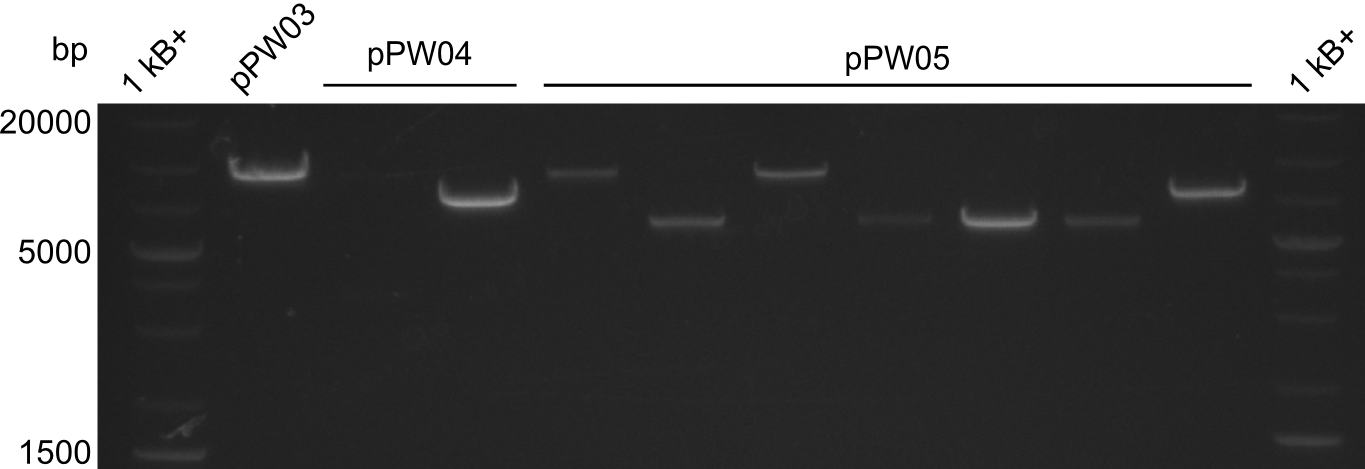
Validation
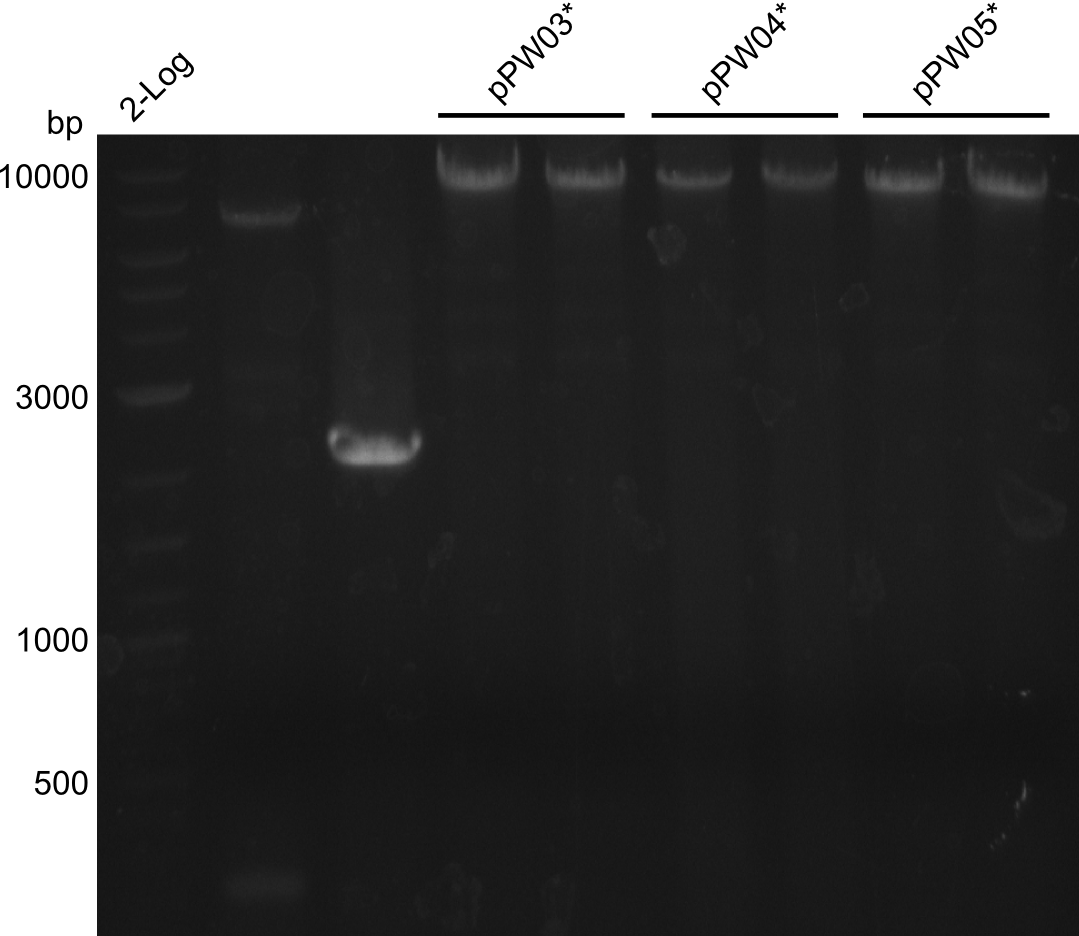
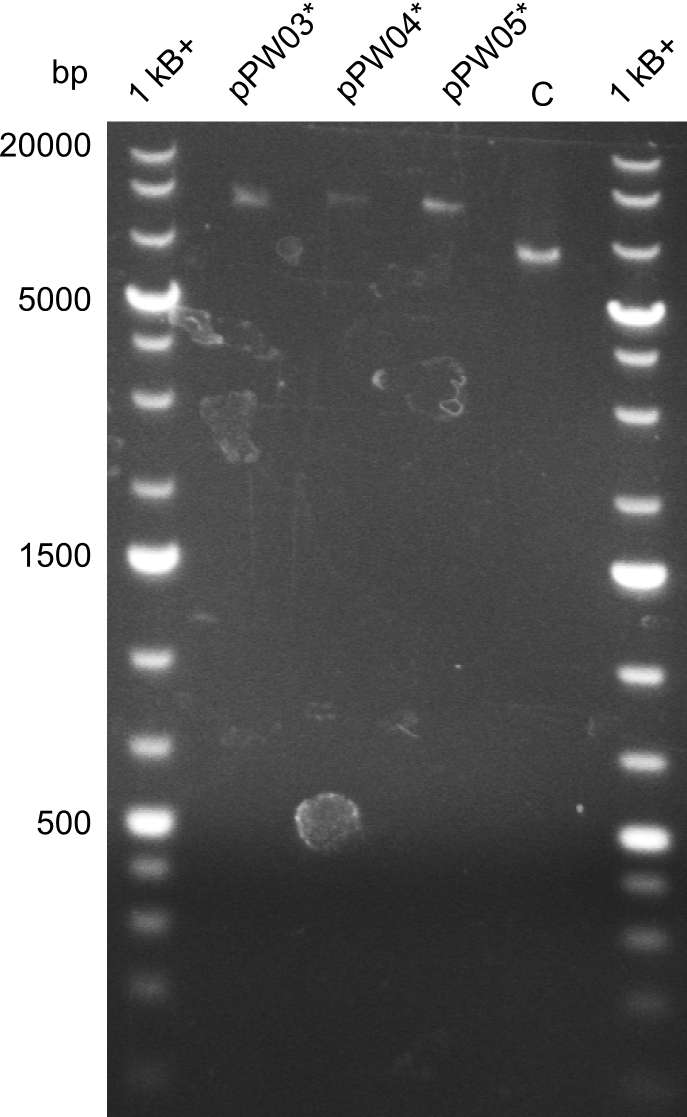
In order to validate colonies that were transformed with the CPEC-Mix, Restriction digest and sequencing was performed. As the unwanted restriction sites should have been mutated in the positive samples, using EcoRI as restriction enzyme should yield a single band at the size of the linearized vector (9049bp for pPW03, 9448bp for pPW04 and 8971bp for pPW05). Restriction digest was conducted with the standard protocol in a 20µl reaction volume.
The image on the lefthand-side shows the gel after 20 minutes at 100V, as this did not lead to good separation, the gel was let run for another 20 minutes. The gel after 40 minutes is displayed on the right side.
The sample for pPW01 and samples 1 and 3 for pPW05 were sent to sequencing as they migrated at the expected size.
Parallel-approach: Reassembly
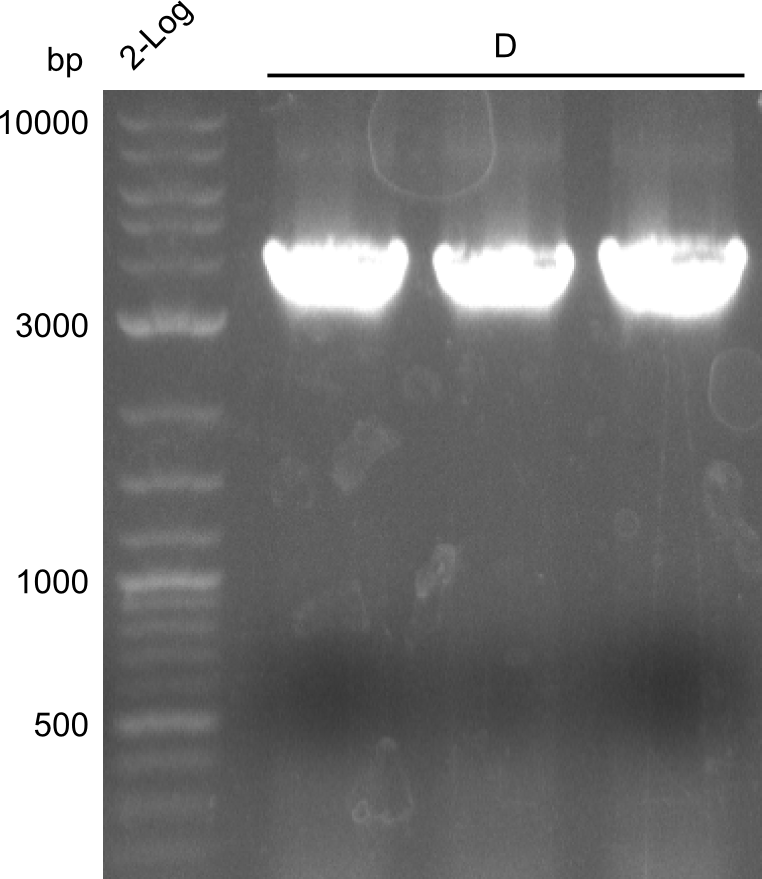
In order to have a second pillar to lean on, we decided to reamplify fragment D (indC) without the two restriction restriction sites. The template for this was created by the Indigoidine-group which had used the CPEC-approach described above.
| what | µl |
|---|---|
| indC* | 0.5 |
| PW15 | 2 |
| PW16 | 2 |
| Phusion Flash 2x Master mix | 10 |
| ddH20 | 5.5 |
| Cycles | temperature [°C] | Time [s] |
|---|---|---|
| 1 | 98 | 5 |
| 12 | 98 | 5 |
| 61 | 5 | |
| 72 | 100 | |
| 23 | 98 | 5 |
| 58 | 10 | |
| 72 | 100 | |
| 1 | 72 | 600 |
| 1 | 10 | inf |
We obtained (as expected) clearly visible, bright bands at the expected fragment size (~4000bp)
 "
"