Team:Leicester/Project
From 2013.igem.org
| Home | Team | Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
Go Leic-styRRRene!
Polystyrene is a multifaceted material - light, largely chemically and biologically inert, and easily shaped, but also a highly visible pollutant that locks up valuable hydrocarbons. In 2013 the Leicester iGEM team are retaining their focus on polystyrene but will diversify our approach. We have three streams involving recycling, re-using and re-purposing polystyrene.
Recycling – This stream involves building on the work of the 2012 team in environmental prospecting for genes involved in polystyrene degradation. Currently we have isolated organisms from environmentally exposed polystyrene, and are progressing in our aim of adapting the toluene degradation pathway of Pseudomonas species.
Re-using - Consumer 3D printers are now a reality and use a variety of thermoplastics (PS and ABS, for example). While the technology is constantly finding new uses, most involve using virgin plastics, which is not good for the environment. Recycled polystyrene can be used for 3D printing, but has an additional advantage as it is soluble in limonene (an environmentally friendly solvent), but ABS is not. Complex 3D printed shapes require removable support structures - one solution is to print the supports using PS and the object in ABS, with the PS being removed by limonene dissolution. We are adapting limonene biosynthesis biobricks developed in previous iGEM competitions to enable genetically engineered machine biological "finishing" of 3D printed objects.
Re-purposing (DNA!) - Expanded polystyrene (EPS) is a great insulator, used by the construction industry to make our homes warmer, using less energy. EPS insulation is required to be flame retardant, which is currently achieved by incorporating environmentally polluting halogenated hydrocarbons. Recently DNA has been shown to be an effective flame retardant, but is expensive to produce. We are building a genetically engineered machine with inducible endoreplication (over-replication of DNA). This should yield DNA cheap enough to burn and when added to EPS make it flame retardant and environmentally friendly.
Recycling
The first of the three streams was focused into amplifying the genes of the tod operon present in the Pseudomonas putida F1 strain. The tod operon is known to degrade toluene. Toluene is degraded by mediation of a dioxygenase. The toluene structure is similar to the styrene monomer structure of polystyrene. The team’s objective was to amplify and modify the dioxygenase genes responsible for toluene degradation so it would accept polystyrene.
The todC1C2BA genes present in the tod operon encode the toluene dioxygenase (TDO), which is capable of oxidizing over a 100 substrates. The catalytic oxygenase is encoded by todC1C2. I was determined the presence of a gap directly into the active site, that could be widen to fit polystyrene. Modelling carried out by the Leicester 2012 iGEM team using the programme Pymol, they modified amino acids present in the gap to amino acids with smaller side chains to widen the gap that could allow polystyrene into the active site. The residues that they modified were Met220 -> Ala220, Val421 -> Ala421, Tyr422 -> Leu422 and Tyr266 -> Val266.
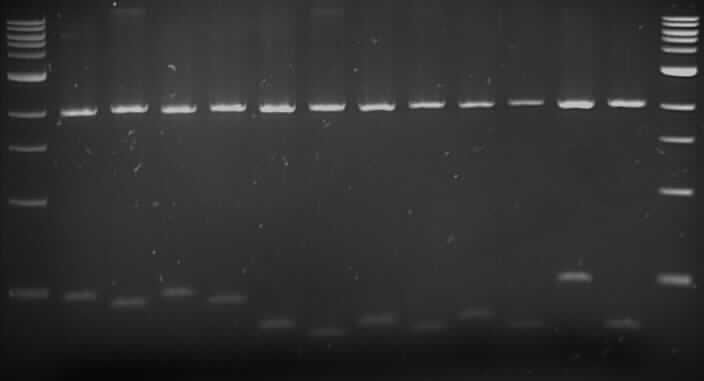
We received the Pseudomonas Putida strain F1. We grew the bacteria overnight on plates, using PCR we amplified the genes tod C, X, F, G and B of the tod operon, also the 16s gene was amplified to confirm that we had the right strain of P.putida. The amplified 16s was sent to be sequenced and sequence was aligned using BLAST to the genomic sequence (NC_009512) of Pseudomonas putida. The alignment confirmed that we had the F1 strain. We proceed to carry out a fusion PCR for the tod genes F, X and B. We made 3 tod genes into biobricks: tod X, F and B. The genes were ligated to the iGEM supplied backbone (pSB1C3), performed a restriction digest on both. The ligations were transformed into DH5α. After the colonies were picked and put on for an overnight culture, the isolated plasmids were sent to be sequences. Sequencing confirmed that the genes were cloned into the backbone and the restrictions sites required by iGEM were present. Further double digestions were carried out to confirm that no mutations occurred.
Gel Order:
F8 (E/P), F8 (X/S); F10 (E/P), F10 (X/S); F12 (E/P), F12 (X/S); B4 (E/P), B4(X/S); B5 (E/P), B5 (X/S); B9 (E/P), B9 (X/S); B12 (E/P), B12 (X/S).
 "
"