The MaGellin Software Package
Open Source Automated, Quantifiable DNA Analysis for our Assay
For a detailed, graphical explanation of the MaGellin work flow, please download the MaGellin Workflow Specifications Sheet, which includes all of the steps in the MaGellin workflow.
The MaGellin Software Package is a MATLAB script that uses computer vision algorithms to calculate the location and intensity of DNA bands. It also has a bioinformatics module to compare the lengths of bands with the expected lengths, based on the methylation sensitivity of the enzymes and the sequence of the plasmid. So, this program is more than another gel quantifier - it interprets the biological meaning of the band lengths and returns experimentally relevant analyses. The MaGellin Software Package is crucial for our workflow because it allows for clear input/output. Since our restriction digests always yield bands in predictable locations, user bias is eliminated and data sets are standardized across trials and labs, accelerating the pace of discovery.
Protocol
1. Upload gel picture from restriction enzyme digest. Fill out all relevant information on the graphical user interface. Enter plasmid and target sequences and select restriction enzymes used. Enter descriptive names for gel and lanes if desired.
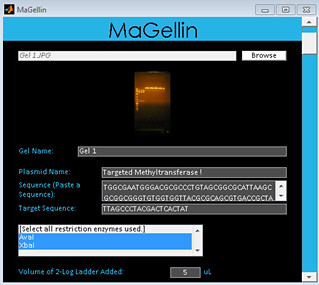
2. Press the Analyze button.
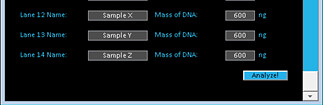
3. The Magellin Software Package will calculate the intensity and position of each band and produce a graph. Save your results.
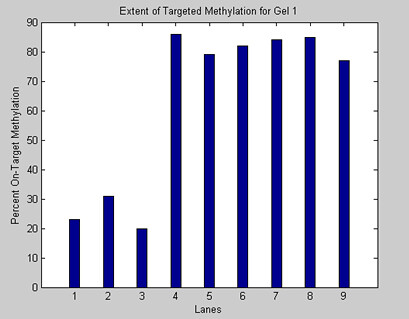 >
>
4. Collaborate with your fellow scientists by sharing your results on SkyDrive.

Next→
1. Upload gel picture from restriction enzyme digest. Fill out all relevant information on the graphical user interface. Enter plasmid and target sequences and select restriction enzymes used. Enter descriptive names for gel and lanes if desired.

2. Press the Analyze button.

3. The Magellin Software Package will calculate the intensity and position of each band and produce a graph. Save your results.
 >
>4. Collaborate with your fellow scientists by sharing your results on SkyDrive.

 "
"

