Team:SDU-Denmark/Tour34
From 2013.igem.org
Heidi.Wille (Talk | contribs) |
|||
| (4 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
</p> | </p> | ||
| - | < | + | |
| - | + | <img class="alignRight" src="https://static.igem.org/mediawiki/2013/4/4e/SDU2013_Safety_1.jpg" style="width:350px" /> | |
| - | + | ||
<p> | <p> | ||
| Line 72: | Line 72: | ||
<span class="intro">Vectors</span><br> | <span class="intro">Vectors</span><br> | ||
| - | |||
<a class="dialogLink" href="http://parts.igem.org/Part:pSB1A3">pSB1A3:</a> An iGEM plasmid backbone carrying an ampicillin resistance gene<br> | <a class="dialogLink" href="http://parts.igem.org/Part:pSB1A3">pSB1A3:</a> An iGEM plasmid backbone carrying an ampicillin resistance gene<br> | ||
| + | <a class="dialogLink" href="http://parts.igem.org/Part:pSB1C3">pSB1C3:</a> An iGEM plasmid backbone carrying a chloramphenicol resistance gene<br> | ||
<a class="dialogLink" href="http://parts.igem.org/Part:pSB1K3">pSB1K3:</a> An iGEM plasmid backbone carrying a kanamycin resistance gene <br> | <a class="dialogLink" href="http://parts.igem.org/Part:pSB1K3">pSB1K3:</a> An iGEM plasmid backbone carrying a kanamycin resistance gene <br> | ||
| Line 81: | Line 81: | ||
<a class="dialogLink" href="http://parts.igem.org/Part:BBa_K1088000">BBa_K1088000</a><br> | <a class="dialogLink" href="http://parts.igem.org/Part:BBa_K1088000">BBa_K1088000</a><br> | ||
| - | Donor: B. | + | Donor: <span class="specialWord">B. Subtilis</span> #168, biosafety level 1<br> |
Enzyme: <span class="tooltipLink">Dxs</span> <span class="tooltip"><span class="tooltipHeader">Dxs</span>1-deoxyxylulose-5-phosphate synthase</span><br> | Enzyme: <span class="tooltipLink">Dxs</span> <span class="tooltip"><span class="tooltipHeader">Dxs</span>1-deoxyxylulose-5-phosphate synthase</span><br> | ||
Possible safety issues: None<br> | Possible safety issues: None<br> | ||
| Line 130: | Line 130: | ||
<p> | <p> | ||
| - | A second possibility is to make the bacteria dependant on a substance only available in the lab: an essential gene could be deleted from the chromosome and inserted on our plasmids. This gene would then be put under the regulation of an inducible promoter. For this to be effective, the substance must be exclusive to the lab environment. | + | <span class="intro">A second possibility</span> is to make the bacteria dependant on a substance only available in the lab: an essential gene could be deleted from the chromosome and inserted on our plasmids. This gene would then be put under the regulation of an inducible promoter. For this to be effective, the substance must be exclusive to the lab environment. |
</p><p> | </p><p> | ||
Latest revision as of 01:12, 29 October 2013
Safety
Prevention prevails
Link to our approved safety formDealing with Genetically Modified Organisms (GMOs) requires an awareness of safety issues. This especially applies to the field of synthetic biology with increasing ease of access to information, lab equipment, and gene technologies. First of all, the safety of public health and the environment must be addressed, but secondly the safety of the researchers must indisputably be considered, too. This applies mainly to the biological agents and products in the lab, but also to the chemical agents and anything else that can pose a potential risk to either group.

In Denmark, a national decree on work involving GMOs (”Bekendtgørelse om godkendelse af produktion med genetisk modificerede mikroorganismer”) addresses the issues of biosafety and provides guidelines for the design of the lab and the handling of the microorganisms. The Institute of Biochemistry and Molecular Biology (BMB), and hence our team, follows this decree. With regards to class 1 labs, the Danish decree resembles the “Laboratory biosafety manual” from the World Health organization to a wide extent.
According to the Danish decree, an assessment of the possible dangers to human safety and health, as well as to the outer environment of the biological systems used and produced in the lab, must be done before the project is executed in the lab. Additionally, the lab classification must be validated by the Danish Working Environment Authority. You will find the risk assessment further down the page. With regards to the lab classification and validation, the lab is well established and it’s classification has been previously validated.
All team members active in the wet lab received a mandatory lab safety course held by Simon Rose (a representative of the work environment group at BMB) before embarking on the wet-lab adventure. From him, we also got a handbook in lab safety. The work environment group at BMB is the equivalent of a biosafety committee. The group concerns itself with the general work environment for the employees and, as a part of this, with the safety of the lab. With only three members of our team having done projects in a class 1 GMO lab prior to this summer, safety concerns could be raised. However, since our team members completed the lab safety course before embarking on lab work, our project was approved and good to go.
Moreover, the team members who participated in team DTU-Denmark’s workshop on synthetic biology did a separate electronic lab safety course before we were allowed to enter their lab. Both lab safety courses covered how to work with genetically modified microorganisms (good laboratory practice), how to handle waste, use of safety devices and what to do in case of an emergency. Researchers Safety During our project our lab team members came into contact with several potentially harmful chemical agents, such as ethidium bromide, DMSO (dimethyl sulfoxide), and antibiotics. When appropriate, gloves were worn and reagents handled in a fume hood. GMOs were always handled wearing gloves. Furthermore, lab members washed their hands after wearing gloves and before exiting the lab. Also, team members wore lab coats when working in the lab, and took them off before exiting the lab area.
A UV board was used for viewing bands in agarose gels. UV rays are also a minor source of potential danger as high rates of radiation are carcinogenic. Yet, the dose from the table is quite small, and both a fixed screen and a wearable facial screen are provided in the room. Also, it is emphasized not to leave the UV board on longer than necessary.
According to Simon Rose, the lab safety committee representative, there should be no significant risk to the researchers when good laboratory practice and the lab’s safety protocol was followed. All organisms we worked with are non-pathogenic, which further added to the safety of the researchers.
Public safety and the environment
With regards to public and environmental safety, our project is only for “contained use”. We are planning on containing our modified bacteria in a closed environment in order to harvest the produced rubber, and all waste from the lab is either autoclaved or inactivated with iodoform. Therefore any public and environmental safety issues would arise from accidents, where the bacteria are released in nature. Good laboratory conduct ought to minimize this risk. However, should our bacteria be released into nature by the incautious or unscrupulous the implications must be considered.A small, but potential danger to the public from our project is the possible outlet of antibiotics which could result in some pathogenic bacteria becoming resistant. We have minimized the range of antibiotics used in our project to chloramphenicol, kanamycin, and ampicillin. Combined with the fact that the strains of E. coli we used carry no other antibiotics resistance, the risk of them surviving in nature and passing on the plasmid is increasingly small. If the bacteria would survive in nature, the most likely case is that the plasmid containing dxs will be lost, since these genes slow down growth and become redundant in a chloramphenicol free environment. Similarly, in a kanamycin free environment, the plasmid containing the prenyltransferase would likely be lost. Otherwise bacteria will conceivably die from poisoning by rubber before sharing any DNA, as we assume that the produced rubber in the bacterial cytoplasm will interrupt it’s natural functions. If the bacteria were to survive in nature containing the plasmid, it would most likely be outcompeted by the naturally occurring and better adapted E. coli strains.
Should a large amount of bacteria be released, the odds of the above mentioned scenarios happening obviously increases, but only fractionally. Another concern with a large-scale release is the possible amount of rubber waste. When a rubber tree dies and the rubber leaks out, plants can still occupy the area; it is possible that this is by plants specialized to live in areas with regular rubber leaks. We cannot know if plants in others area can survive when rubber particles are present, and hence whether this could damage a local environment.
Furthermore, the representative of the work environment group pointed out that E. coli could be used to contaminate water. However, this was a minimal risk, and smaller than the possible contamination by E. coli from other sources.
Risk assessment of individual parts
HostsE. coli K12 MG1655 and KG22, biosafety level 1.
Non-pathogenic strains of E. coli.
Vectors
pSB1A3: An iGEM plasmid backbone carrying an ampicillin resistance gene
pSB1C3: An iGEM plasmid backbone carrying a chloramphenicol resistance gene
pSB1K3: An iGEM plasmid backbone carrying a kanamycin resistance gene
BioBricks
BBa_K1088000
Donor: B. Subtilis #168, biosafety level 1
Enzyme: Dxs Dxs1-deoxyxylulose-5-phosphate synthase
Possible safety issues: None
BBa_K1088003
Donor: Hevea brasiliensis, biosafety level 1
Enzyme: HRT2 - Prenyltransferase
Possible safety issues: None
BBa_K1088004
Donor: E. coli K12 MG1655, biosafety level 1
Enzyme: IspG IspG(E)-4-hydroxy-3-methylbut-2-mehtyl-D-erythritol-2,4-cyclodiphosphate synthase
Possible safety issues: None
BBa_K1088018
Donor: E. coli K12 MG1655, biosafety level 1
Enzyme: LacI
Possible safety issues: None
Safety improvements
Finally, a couple of possible improvements can be made to enhance the safety of our project.We envision a dual toxin/antitoxin system. This is made possible because of our system design, which has two separate plasmids. The basic idea is to have each plasmid produce a toxin, while the corresponding antitoxin is produced by the other plasmid. In this manner, a bacteria can only survive if both plasmids are present. The risk of horizontal gene transfer (if the bacteria were to escape their physical confines) is drastically reduced with the introduction of a dual toxin/antitoxin system. The likelihood that a wildtype bacteria would take up both plasmids at once is highly unlikely - and the strains would be unable to survive with only a single plasmid. A system resembling this idea has been previously described in the holin/anti-holin biobrick (BBa_K515106/BBa_K515104).
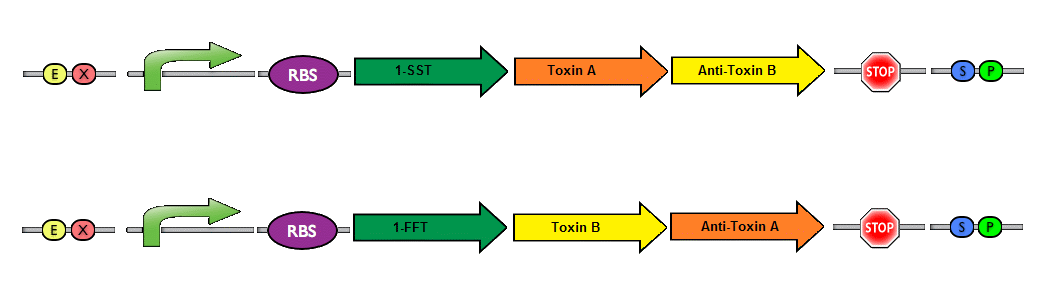 Picture by iGEM team from SDU 2012; Depicting a toxin/antitoxin system
Picture by iGEM team from SDU 2012; Depicting a toxin/antitoxin system
A second possibility is to make the bacteria dependant on a substance only available in the lab: an essential gene could be deleted from the chromosome and inserted on our plasmids. This gene would then be put under the regulation of an inducible promoter. For this to be effective, the substance must be exclusive to the lab environment.
Note from iGEM headquarters:
Safety forms were approved on September 22, 2013 by Evan Appleton.
 "
"