Team:Tsinghua-E/Part2
From 2013.igem.org
| (4 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<style type="text/css"> | <style type="text/css"> | ||
#content{height:1760px;} | #content{height:1760px;} | ||
| - | p{font-size: | + | p{font-size:130%} |
.memberx {position:absolute;top:60px;left:5px;height:215px;width;300px;} | .memberx {position:absolute;top:60px;left:5px;height:215px;width;300px;} | ||
.memberx span{z-index:50;position:relative;width:453px;display:block;top:-215px;left:305px;color:#000000} | .memberx span{z-index:50;position:relative;width:453px;display:block;top:-215px;left:305px;color:#000000} | ||
| Line 23: | Line 23: | ||
</html> | </html> | ||
| - | + | <h2>[[File:4_2s.png|68px|left]] '''Part 2: THU-E Tryptophan-sensor Part'''</h2> | |
| - | [[File: | + | [[File:Plasmid2.png|446px|right]] |
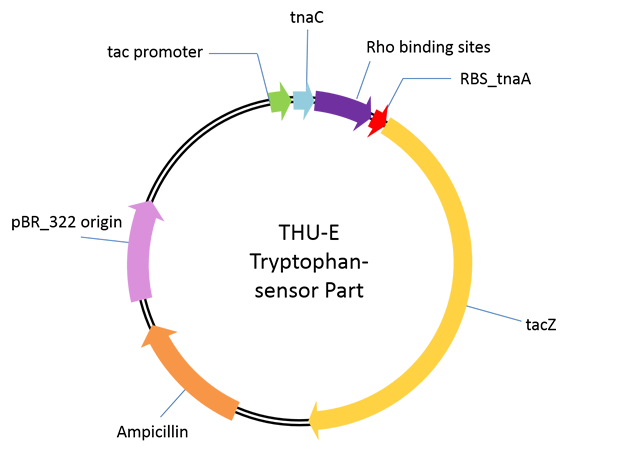
Aplasmid used for performance test of our novel tryptophan sensor. | Aplasmid used for performance test of our novel tryptophan sensor. | ||
| - | The sensor was derived from one regulation sequence upstream of tryptophanase(tnaA) operon in wild type <em>E. Coli</em><html><a href="#_ENREF_1" title="Gong, 2002 #484">3</a></html>. This sequence codesone 24-residue nascent peptide. Following this nascent peptide sequence stands one transcription termination factor (Rho) recognition sites. When certain amount of tryptophan exists, it is recognized by the nascent peptide. This leads to the hindering of TnaC-peptidyl-tRNAPro from being cleaved from ribosome. This peptide-mRNA-ribosome complex blocks Rho factor’s access to its binding site which is just adjacent to termination codon of nascent peptide so that initiate the transcription of downstream sequence. As far as we know, this novel mechanism has not been utilized before as tryptophan sensor. | + | The sensor was derived from one regulation sequence upstream of tryptophanase(tnaA) operon in wild type <em>E. Coli</em><html><a href="#_ENREF_1" title="Gong, 2002 #484">[3]</a></html>. This sequence codesone 24-residue nascent peptide. Following this nascent peptide sequence stands one transcription termination factor (Rho) recognition sites. When certain amount of tryptophan exists, it is recognized by the nascent peptide. This leads to the hindering of TnaC-peptidyl-tRNAPro from being cleaved from ribosome. This peptide-mRNA-ribosome complex blocks Rho factor’s access to its binding site which is just adjacent to termination codon of nascent peptide so that initiate the transcription of downstream sequence. As far as we know, this novel mechanism has not been utilized before as tryptophan sensor. |
As a proof of principle, we cloned beta-lactamase gene <em>lacZ</em> downstream of wild type nascent peptide and Rho interaction sequence. The assembly was cloned between the NcoI and BamHI sites of pTrc99A vector with IPTG induction.By measuring the activity of beta-lactamase activity (the protocol has been described in detail in our note, please refer to it) after induction and 21-hours culture, we obtained our expected tryptophan dependent beta-lactamase activity increase with dynamic range up to 3mM tryptophan. | As a proof of principle, we cloned beta-lactamase gene <em>lacZ</em> downstream of wild type nascent peptide and Rho interaction sequence. The assembly was cloned between the NcoI and BamHI sites of pTrc99A vector with IPTG induction.By measuring the activity of beta-lactamase activity (the protocol has been described in detail in our note, please refer to it) after induction and 21-hours culture, we obtained our expected tryptophan dependent beta-lactamase activity increase with dynamic range up to 3mM tryptophan. | ||
| Line 40: | Line 40: | ||
<br/> | <br/> | ||
| - | + | <br/> | |
| + | <p><font style="font-size: 12px">[3] Gong, F. & Yanofsky, C. Instruction of translating ribosome by nascent peptide. <em>Science</em> 297, 1864-1867, doi:10.1126/science.1073997 (2002).</fontsize></p> | ||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 03:08, 28 September 2013



Part 2: THU-E Tryptophan-sensor Part
Aplasmid used for performance test of our novel tryptophan sensor.
The sensor was derived from one regulation sequence upstream of tryptophanase(tnaA) operon in wild type E. Coli[3]. This sequence codesone 24-residue nascent peptide. Following this nascent peptide sequence stands one transcription termination factor (Rho) recognition sites. When certain amount of tryptophan exists, it is recognized by the nascent peptide. This leads to the hindering of TnaC-peptidyl-tRNAPro from being cleaved from ribosome. This peptide-mRNA-ribosome complex blocks Rho factor’s access to its binding site which is just adjacent to termination codon of nascent peptide so that initiate the transcription of downstream sequence. As far as we know, this novel mechanism has not been utilized before as tryptophan sensor.
As a proof of principle, we cloned beta-lactamase gene lacZ downstream of wild type nascent peptide and Rho interaction sequence. The assembly was cloned between the NcoI and BamHI sites of pTrc99A vector with IPTG induction.By measuring the activity of beta-lactamase activity (the protocol has been described in detail in our note, please refer to it) after induction and 21-hours culture, we obtained our expected tryptophan dependent beta-lactamase activity increase with dynamic range up to 3mM tryptophan.
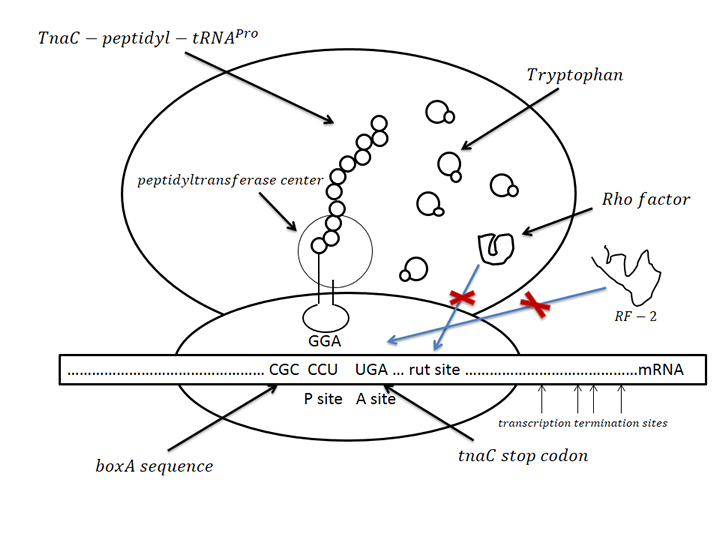
Figure.1 Conception illustration of the working mechanism of novel tryptophan sensor

Figure.2 the performance of tryptophan sensor measured by the dependent relation between tryptophan concentration and beta-lactamase activity
[3] Gong, F. & Yanofsky, C. Instruction of translating ribosome by nascent peptide. Science 297, 1864-1867, doi:10.1126/science.1073997 (2002).
 "
"