Team:Tsinghua-E/Part3
From 2013.igem.org
| (9 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html xmlns="http://www.w3.org/1999/xhtml"> | <html xmlns="http://www.w3.org/1999/xhtml"> | ||
<style type="text/css"> | <style type="text/css"> | ||
| - | #content{height: | + | #content{height:2490px;} |
| - | p{font-size: | + | p{font-size:130%} |
.memberx {position:absolute;top:60px;left:5px;height:215px;width;300px;} | .memberx {position:absolute;top:60px;left:5px;height:215px;width;300px;} | ||
.memberx span{z-index:50;position:relative;width:453px;display:block;top:-215px;left:305px;color:#000000} | .memberx span{z-index:50;position:relative;width:453px;display:block;top:-215px;left:305px;color:#000000} | ||
| Line 17: | Line 17: | ||
</div> | </div> | ||
| - | <div class="tmc" style="height: | + | <div class="tmc" style="height:2375px;"> |
</div> | </div> | ||
| - | <div class="neirong" style="height: | + | <div class="neirong" style="height:2355px;"> |
| - | <h2>Part 3: THU-E | + | |
| - | + | </html> | |
| - | + | ||
| - | + | <h2>[[File:4_3s.png|60px|left]] '''Part 3: THU-E Bitter Defender Part''' </h2> | |
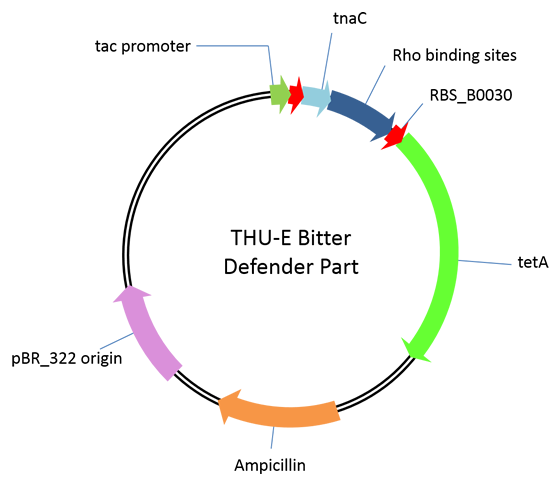
| - | <p align="center"> | + | [[File:Plasmid3.png|446px|right]] |
| - | <img width="446" | + | |
| - | Figure.1 | + | This is a plasmid providing selection pressure for the evolution and enrichment of tryptophan overproduction microorganism phenotype by defending the toxicity of tetracycline. This selection pressure was based on the tryptophan dependent tetracycline antiporterexpression which functioned in tetracycline culture condition.It is achieved by cloning<em> E. Coli</em> tetracycline antiporter gene (<em>tetA</em>) downstream of our previously constructed tryptophan biosensor which wascontrolled by tac promoter in pTrc99A vector. Three different RBS (RBS-ori (RBS upstream of wild type<em>E. Coli</em> tryptophanase(tnaA) operon), RBS-30 (B0030), RBS-32 (B0032)) were placed upstream of <em>tetA</em> gene. |
| - | <p align="center"><img width="446" src="/wiki/images/ | + | |
| - | + | Previously engineered <em>E. coli</em> with different tryptophan productivity (trp000, trp007, trp015 with the overall productivity as 0, 0.07, 0.15g/L, respectively. unpublished data) were used to test the performance of this part. The tryptophan productivity of each strain wasmeasured, andit was clear that transformation of the engineered bitter defender parts didn’t significantly influence original strains’ tryptophan relative productivity according to Figure.3. | |
| - | <p align="center"><img width="446" src="/wiki/images/ | + | |
| - | Figure.3 | + | We thus tested these three parts’ performance by measuring the growth rate of these nine strains (three kinds of strains with different tryptophan productivity carrying three kinds of selection pressure plasmids. They were named after trp000-30, trp000-32, trp000-ori, trp007-30, trp007-32, trp007-ori, trp015-30, trp015-32 and trp015-ori, respectively) in different tetracycline concentration. We interestingly found that strains carrying bitter defender part with RBS B0030 and B0032 showed good tryptophan dependent growth property within the first 15h: |
| + | |||
| + | (1) As the increase of tetracycline, the selection pressure increased and the growth rate of strains decreased. | ||
| + | |||
| + | (2) With increased selection pressure, it could be observed that within time window of 15h, in some selection pressure condition (tetracycline concentration) strains with higher tryptophan productivity showed much higher growth rate, while the growth of tryptophan non producer trp000 nearly inhibited by high concentration of tetracycline. | ||
| + | |||
| + | <html> | ||
| + | |||
| + | <p align="center"> <img width="446" src="/wiki/images/0/00/Part3II.png" /> <br /> | ||
| + | Figure.1 Conception illustration of the working mechanism of bitter defender part <br /></p> | ||
| + | <p align="center"> <img width="446" src="/wiki/images/5/59/Part4I.png" /> <br /> | ||
| + | Figure.2 Tryptophan productivity of previously engineered strains with different productivity carrying three kinds of bitter defender plasmids<br /></p> | ||
| + | <p align="center"> <img width="446" src="/wiki/images/c/cf/Part4Is.png" /> </p> | ||
| + | <p align="center"><img width="446" src="/wiki/images/d/db/Part4Is2.png" /> <br /> | ||
| + | Figure.3 Bitter defender part performance was measured by the dependent relation between the strain growth rate and the intracellular tryptophan productivity in finely tuned M9YE culture condition.Strains carrying bitter defender part with RBS B0030 and B0032 were shown respectively. </p> | ||
| + | |||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 15:13, 27 September 2013



Part 3: THU-E Bitter Defender Part
This is a plasmid providing selection pressure for the evolution and enrichment of tryptophan overproduction microorganism phenotype by defending the toxicity of tetracycline. This selection pressure was based on the tryptophan dependent tetracycline antiporterexpression which functioned in tetracycline culture condition.It is achieved by cloning E. Coli tetracycline antiporter gene (tetA) downstream of our previously constructed tryptophan biosensor which wascontrolled by tac promoter in pTrc99A vector. Three different RBS (RBS-ori (RBS upstream of wild typeE. Coli tryptophanase(tnaA) operon), RBS-30 (B0030), RBS-32 (B0032)) were placed upstream of tetA gene.
Previously engineered E. coli with different tryptophan productivity (trp000, trp007, trp015 with the overall productivity as 0, 0.07, 0.15g/L, respectively. unpublished data) were used to test the performance of this part. The tryptophan productivity of each strain wasmeasured, andit was clear that transformation of the engineered bitter defender parts didn’t significantly influence original strains’ tryptophan relative productivity according to Figure.3.
We thus tested these three parts’ performance by measuring the growth rate of these nine strains (three kinds of strains with different tryptophan productivity carrying three kinds of selection pressure plasmids. They were named after trp000-30, trp000-32, trp000-ori, trp007-30, trp007-32, trp007-ori, trp015-30, trp015-32 and trp015-ori, respectively) in different tetracycline concentration. We interestingly found that strains carrying bitter defender part with RBS B0030 and B0032 showed good tryptophan dependent growth property within the first 15h:
(1) As the increase of tetracycline, the selection pressure increased and the growth rate of strains decreased.
(2) With increased selection pressure, it could be observed that within time window of 15h, in some selection pressure condition (tetracycline concentration) strains with higher tryptophan productivity showed much higher growth rate, while the growth of tryptophan non producer trp000 nearly inhibited by high concentration of tetracycline.

Figure.1 Conception illustration of the working mechanism of bitter defender part

Figure.2 Tryptophan productivity of previously engineered strains with different productivity carrying three kinds of bitter defender plasmids
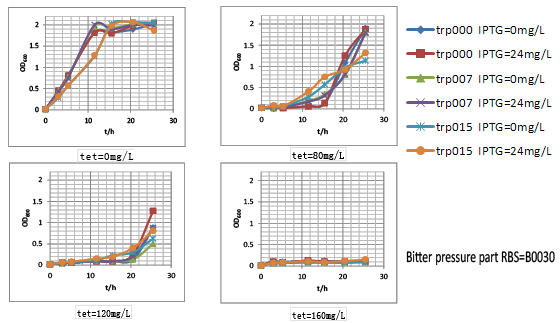

Figure.3 Bitter defender part performance was measured by the dependent relation between the strain growth rate and the intracellular tryptophan productivity in finely tuned M9YE culture condition.Strains carrying bitter defender part with RBS B0030 and B0032 were shown respectively.
 "
"