Team:Tsinghua-E/Project
From 2013.igem.org
| Line 2: | Line 2: | ||
<html xmlns="http://www.w3.org/1999/xhtml"> | <html xmlns="http://www.w3.org/1999/xhtml"> | ||
<style type="text/css"> | <style type="text/css"> | ||
| - | #content{height: | + | #content{height:1800px;} |
p{font-size:120%} | p{font-size:120%} | ||
.memberx {position:absolute;top:60px;left:5px;height:215px;width;300px;} | .memberx {position:absolute;top:60px;left:5px;height:215px;width;300px;} | ||
| Line 16: | Line 16: | ||
</div> | </div> | ||
| - | <div class="tmc" style="height: | + | <div class="tmc" style="height:1685px;"> |
</div> | </div> | ||
| - | <div class="neirong" style="height: | + | <div class="neirong" style="height:1665px;"> |
<h2>Overview</h2> | <h2>Overview</h2> | ||
<h3 align="center"><strong>Darwinian evolution for microbial cell factory:</strong><br /> | <h3 align="center"><strong>Darwinian evolution for microbial cell factory:</strong><br /> | ||
Revision as of 15:40, 22 September 2013



Overview
Darwinian evolution for microbial cell factory:
in vivo evolution engineering towards tryptophan-overproduction superbug
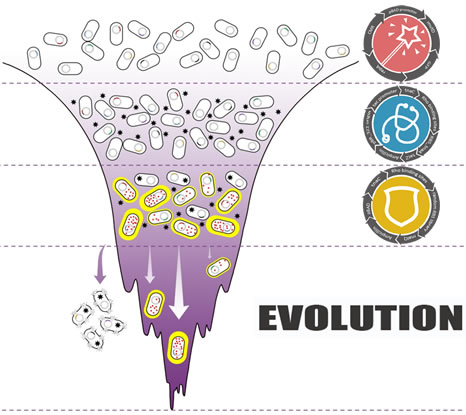
Darwinian evolution shows great power in creating incrediblebiological functionin amazing speed.Inspired by this, our team aimed atcreating novelfast and irrational microbial cell factory by simulating natural Darwinian evolution process. With tryptophan as target product, a novel tryptophan biosensorutilizing translating ribosome mechanismwas firstly developed as the foundation for tryptophan productivity and selection pressure switch module. We further constructedthis tryptophan overproduction selection gene circuit couplingwith in vivomutationmachine (mutator gene of mutD). By fine-tuning the selection conditions, our selection circuit showed good tryptophan dependent growth property, which provides the foundation for further evolution.As a preliminary result of this project, we successfully evolved an ancestor with zero productivity to a high-tryptophan producer only after several rounds of evolution.
Protocol
(1) Strains:
BL21(DE3)(mut-bitter30(+))/pSC101wt-ori-mutD-sfGFP/pTrc99A-trpss-30-tetA BL21(DE3)(mut-bitter32(+))/pSC101wt-ori-mutD-sfGFP/pTrc99A-trpss-32-tetA Negative control: BL21(DE3) pSC101wt-ori-mutD-sfGFP/pTrc99A-trpss-30-tetA BL21(DE3) pSC101wt-ori-mutD-sfGFP/pTrc99A-trpss-32-tetA Each evolution was conducted in three parallel with only one parallel of negative control.
(2) Genome mutation
Colonies were picked up from agar plate to culture in 5mL LB medium in a 15mL centrifuge tube with 50mg/L ampicillin and chloromycetin for overnight. The seed culture was added into the fresh 10mL LB medium with the same antibiotics in 50mL flask by volumn ratio of 1%. 1g/L arabinose was added during the component stage (OD600= 0.6-0.8). The culture was maintained at 37℃ for 24h.
(3) Evolution
The culture was centrifuged and resuspended by 2mL M9YE medium and cultured in 20mL M9YE medium with 50mg/L ampicillin, chloromycetin, 24mg/L IPTG and certain concentration of tetracycline (initial OD600 was about 0.02). OD600 of the culture was measured every two hour and transferred to the next M9YE medium culture with higher tetracycline concentration with OD600 within 0.3-0.35. For every round, 1mL culture was mixed with 1mL 50% glycerol and stored. One fraction of culture was cultured on agar plate to obtain single colony for further analysis. The remaining culture was continuouslymaintain in culture condition to stable stage for tryptophan HPLC analysis.
The evolution for each condition was conducted for three rounds. The concentration of tetracycline of the culture during evolution process was set as below.
Strains |
1th round |
2th round |
3th round |
Mut-30(+) |
80mg/L |
120mg/L |
160mg/L |
Mut-32(+) |
120mg/L |
160mg/L |
200mg/L |
Table tetracycline concentration increase during the evolution process
 "
"