Team:UCLA/Modeling/Analysis
From 2013.igem.org
Anuvedverma (Talk | contribs) |
Michaelc1618 (Talk | contribs) |
||
| (8 intermediate revisions not shown) | |||
| Line 37: | Line 37: | ||
<li><div id="spec"><a href="/Team:UCLA/Modeling"><font color="black">MODELING</font></a></div></li> | <li><div id="spec"><a href="/Team:UCLA/Modeling"><font color="black">MODELING</font></a></div></li> | ||
<li><a href="/Team:UCLA/HumanPractices">HUMAN PRACTICES</a></li> | <li><a href="/Team:UCLA/HumanPractices">HUMAN PRACTICES</a></li> | ||
| - | <li><a href="/Team:UCLA/Notebook">NOTEBOOK</a></li> | + | <li><a href="/Team:UCLA/Notebook/Biobrick">NOTEBOOK</a></li> |
<li><a href="/Team:UCLA/Safety">SAFETY</a></li> | <li><a href="/Team:UCLA/Safety">SAFETY</a></li> | ||
<li><a href="/Team:UCLA/Attributions">ATTRIBUTIONS</a></li> | <li><a href="/Team:UCLA/Attributions">ATTRIBUTIONS</a></li> | ||
| Line 63: | Line 63: | ||
== '''Analysis of Protein''' == | == '''Analysis of Protein''' == | ||
| - | + | ===Software=== | |
| - | + | ||
| + | <div> | ||
| + | Analysis of the Mtd protein was conducted using our own 'Protein Calculator' software alongside PyMOL. The Protein Calculator allows users to see the potential effects of polymorphic nucleotides in their DNA library on the protein level. The PyMOL software allows users to upload, visualize, and modify molecules of interest in 3D. Together, these two programs allowed our team to locate variable amino acids in the Mtd, as well as predict the effects on binding affinity that the polymorphic nucleotides can have on the resulting protein. | ||
| - | |||
| - | + | Instructions to download and use our program are as follows. (<b>Disclaimer!</b> There are still several bugs in the Protein Calculator program that are still in the process of being fixed. The dropbox download link below will be updated with newer versions as the fixes are made.) | |
| - | + | 1) Click the link dropbox link below and download the files. | |
| + | [[File:Walkthrough-1.png|thumb|left|Step 1.|800px]] | ||
| - | + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | |
| + | 2) Your computer may display some type of warning- ignore the warning to download the program. | ||
| - | + | [[File:Walkthrough-2.png|thumb|left|Step 2.|800px]] | |
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | 3) Extract the zip file into the directory/location of your choice. | ||
| + | |||
| + | [[File:Walkthrough-3.png|thumb|left|Step 3.|800px]] | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | 4) Open Sequence.txt to enter the DNA sequence you wish to analyze. Be sure to keep Sequence.txt in the same directory as Protein Calculator.exe. <b>Important: </b> enter the sequence such that it is within an appropriate reading frame (number of base pairs must be multiple of 3), the nucleotides are in all capital letters, and entered in the 5' -> 3' direction. | ||
| + | |||
| + | [[File:Walkthrough-4.png|thumb|left|Step 4.|800px]] | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | 5) Once Sequence.txt contains your DNA sequence in the appropriate format, open Protein Calculator.exe. Another warning may appear- click 'Run Anyway' to run the program. | ||
| + | |||
| + | [[File:Walkthrough-5.png|thumb|left|Step 5.|800px]] | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
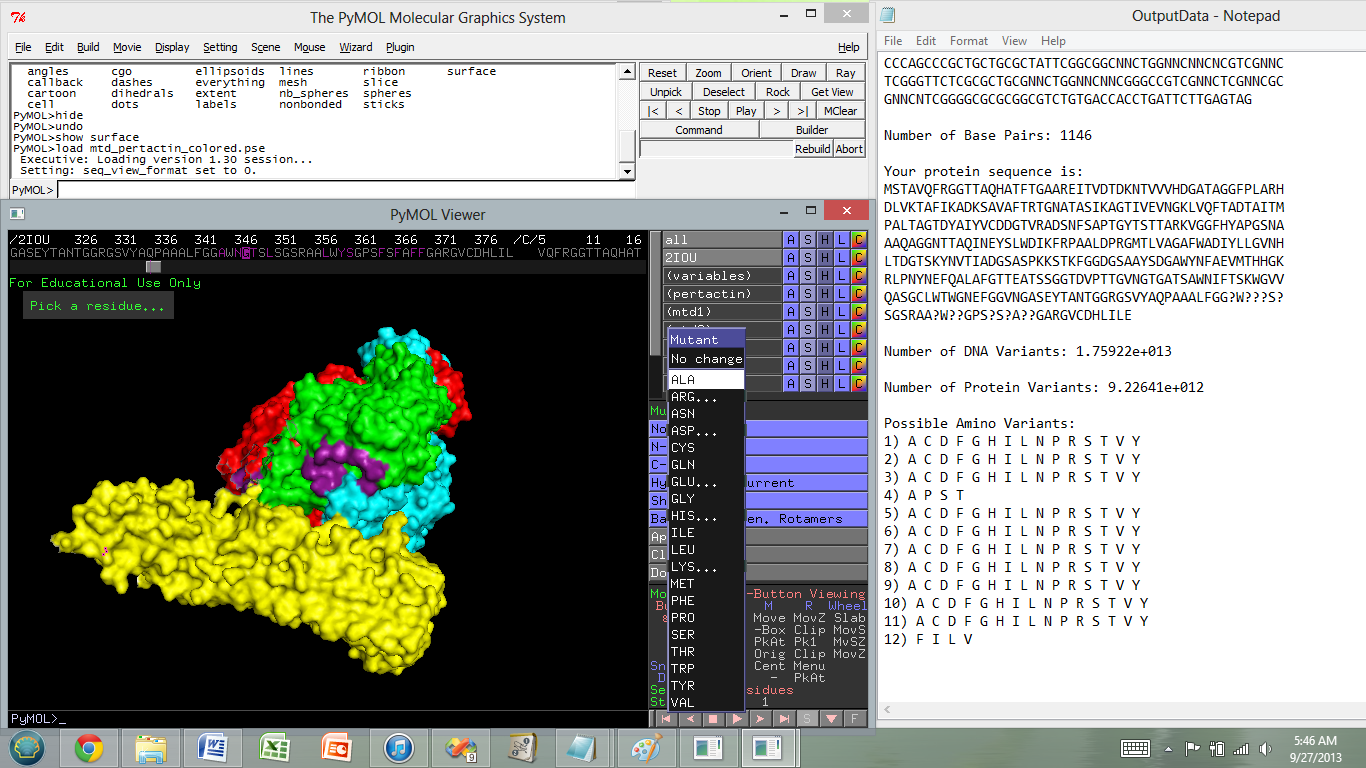
| + | 6) A cmd window will open with instructions. A new .txt file should appear in the same directory as Protein Calculator.exe called OutputData.txt. This text file contains information about your polymorphic DNA sequence library. | ||
| + | |||
| + | [[File:Walkthrough-6.png|thumb|left|Step 6.|800px]] | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | 7) Open OutputData.txt to view resulting data. Before closing the program, copy this data onto a separate file- all text will be deleted upon closing the program (by entering '0' in the cmd window). | ||
| + | |||
| + | [[File:Walkthrough-7.png|thumb|left|Step 7.|800px]] | ||
| + | |||
| + | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
| + | 8) This data can now be used to accurately locate and modify variable regions in the protein of interest. | ||
| + | |||
| + | [[File:Walkthrough-8.png|thumb|left|Step 8.|800px]] | ||
| + | |||
| + | </div> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | https://www.dropbox.com/s/bai70ev5oqksj98/Protein%20Calculator.zip | ||
| + | |||
| + | <!-- | ||
<div id="reference"> | <div id="reference"> | ||
<b>REFERENCES</b> | <b>REFERENCES</b> | ||
| Line 85: | Line 154: | ||
</div> | </div> | ||
| + | --> | ||
Latest revision as of 23:28, 27 September 2013
Analysis of Protein
Software
Analysis of the Mtd protein was conducted using our own 'Protein Calculator' software alongside PyMOL. The Protein Calculator allows users to see the potential effects of polymorphic nucleotides in their DNA library on the protein level. The PyMOL software allows users to upload, visualize, and modify molecules of interest in 3D. Together, these two programs allowed our team to locate variable amino acids in the Mtd, as well as predict the effects on binding affinity that the polymorphic nucleotides can have on the resulting protein.
Instructions to download and use our program are as follows. (Disclaimer! There are still several bugs in the Protein Calculator program that are still in the process of being fixed. The dropbox download link below will be updated with newer versions as the fixes are made.)
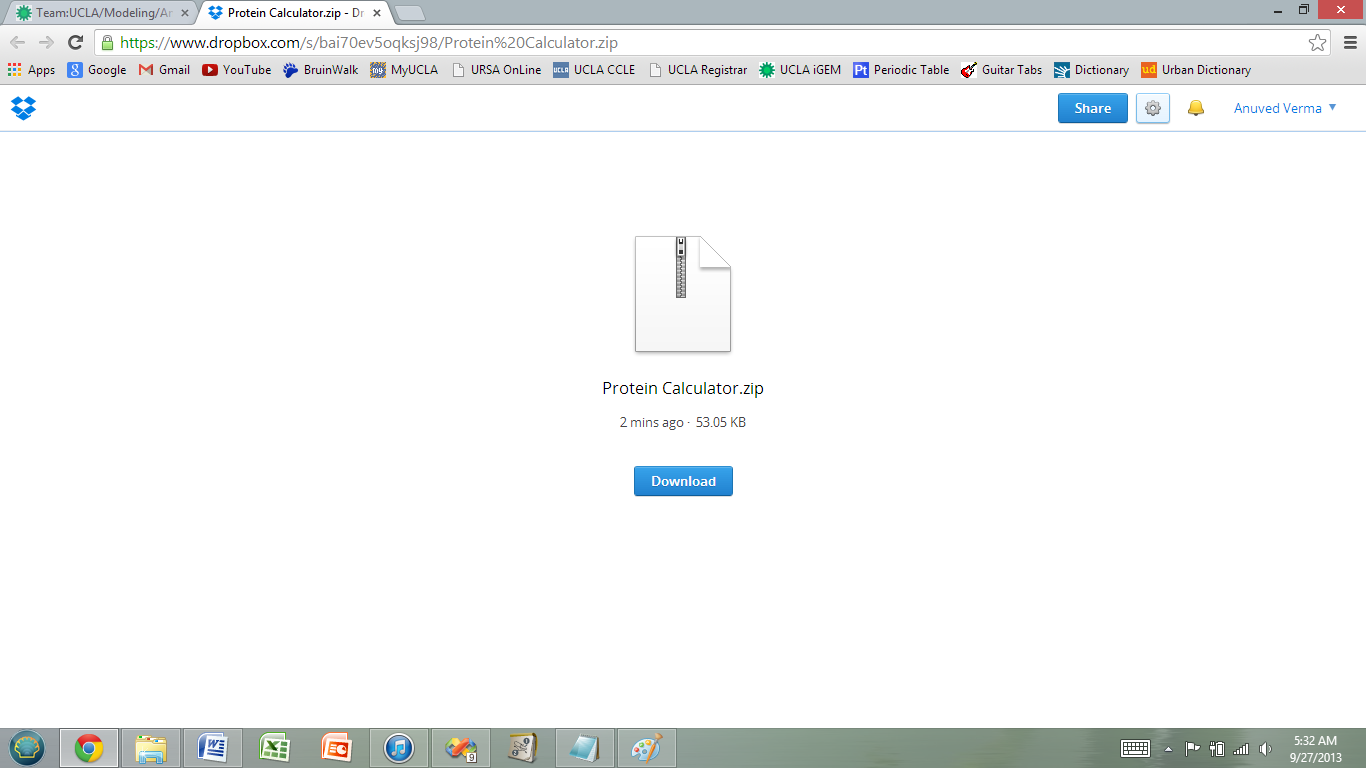
1) Click the link dropbox link below and download the files.
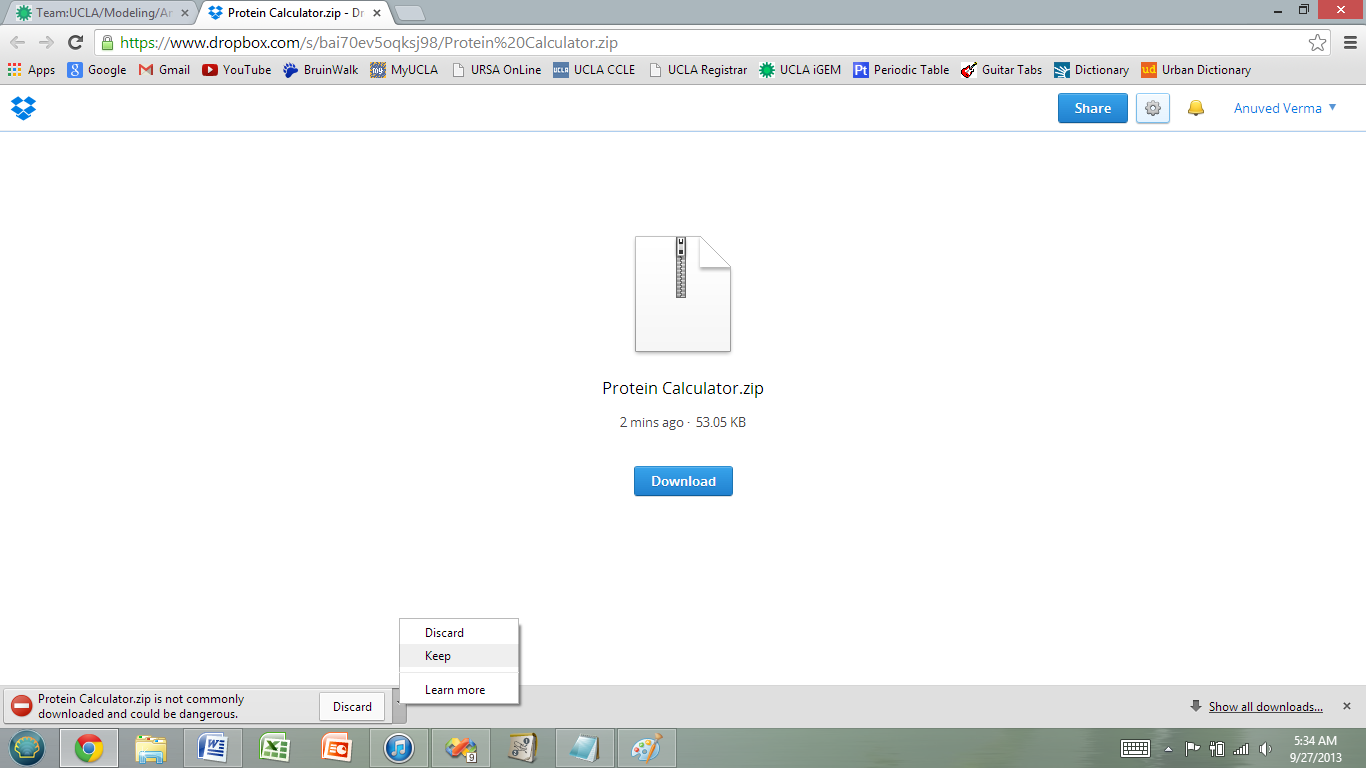
2) Your computer may display some type of warning- ignore the warning to download the program.
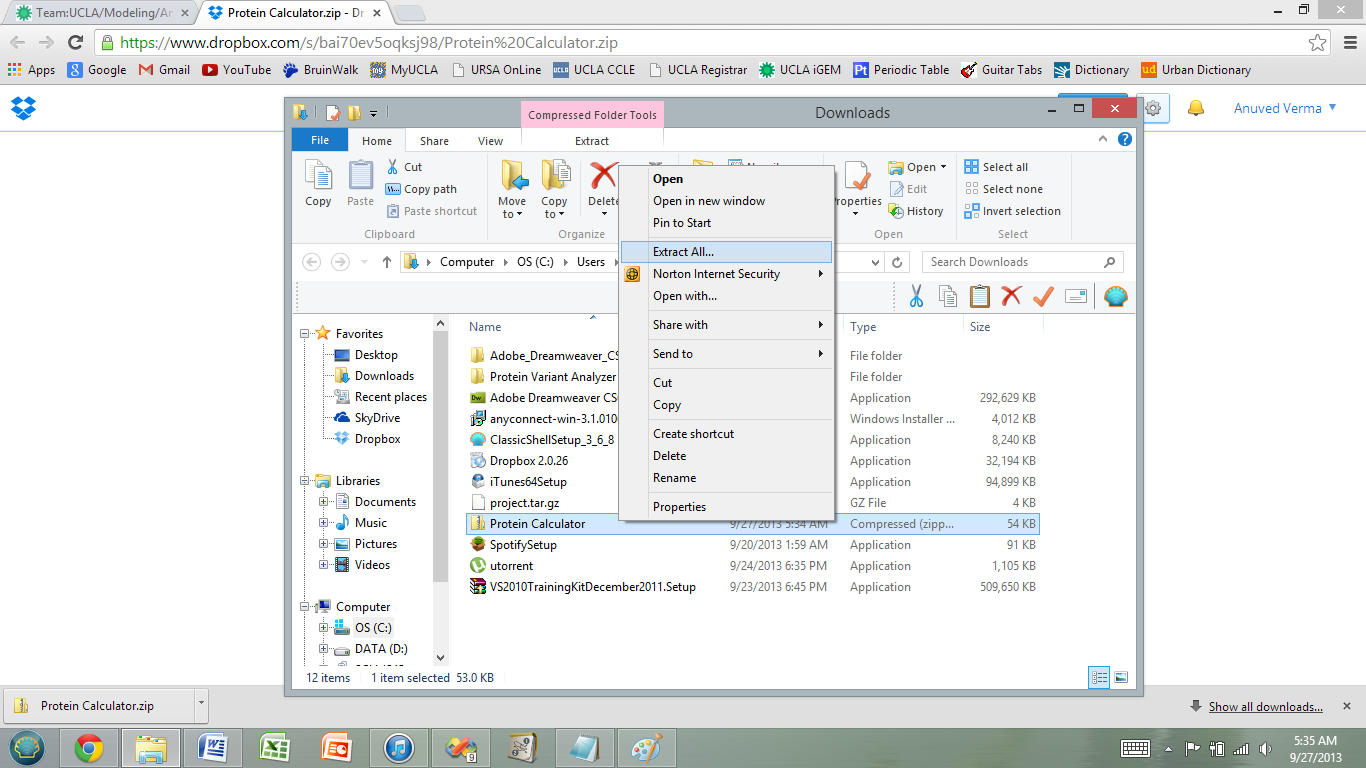
3) Extract the zip file into the directory/location of your choice.
4) Open Sequence.txt to enter the DNA sequence you wish to analyze. Be sure to keep Sequence.txt in the same directory as Protein Calculator.exe. Important: enter the sequence such that it is within an appropriate reading frame (number of base pairs must be multiple of 3), the nucleotides are in all capital letters, and entered in the 5' -> 3' direction.
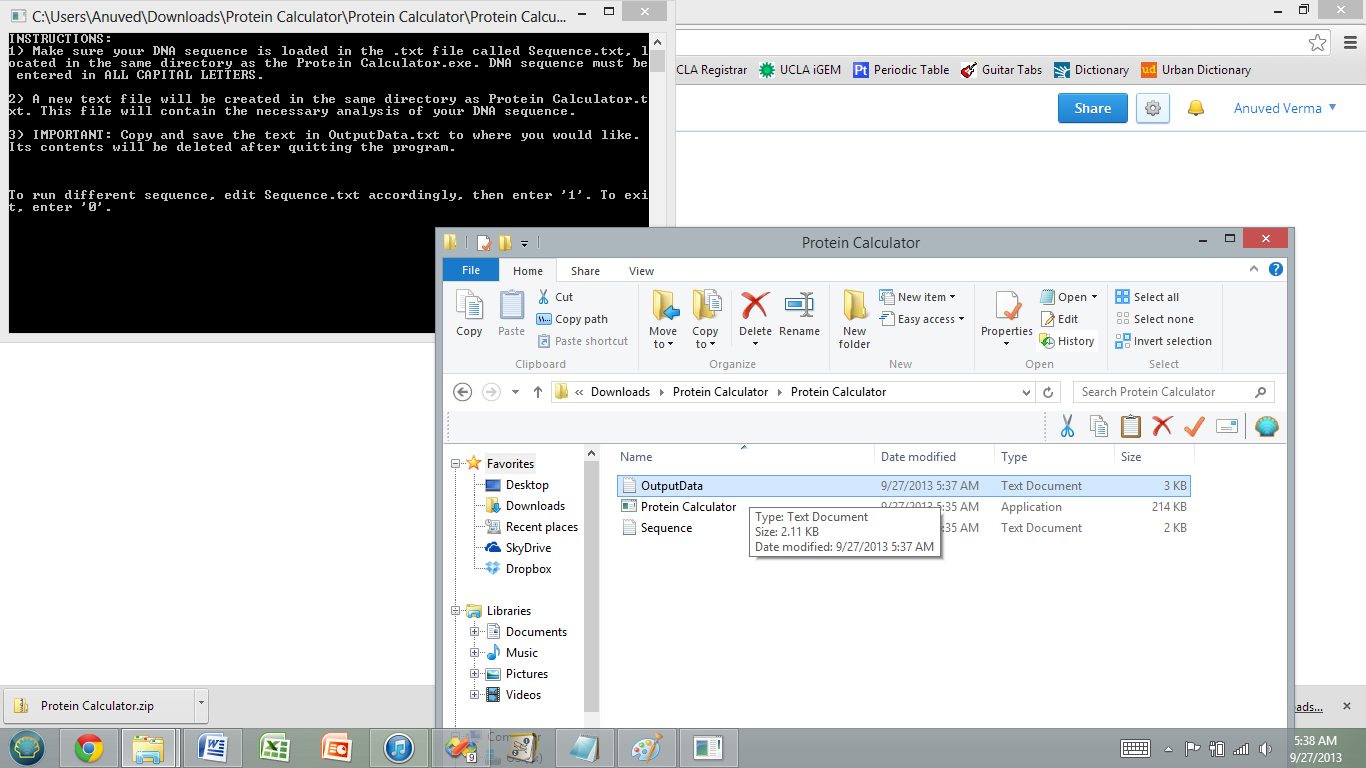
5) Once Sequence.txt contains your DNA sequence in the appropriate format, open Protein Calculator.exe. Another warning may appear- click 'Run Anyway' to run the program.
6) A cmd window will open with instructions. A new .txt file should appear in the same directory as Protein Calculator.exe called OutputData.txt. This text file contains information about your polymorphic DNA sequence library.
7) Open OutputData.txt to view resulting data. Before closing the program, copy this data onto a separate file- all text will be deleted upon closing the program (by entering '0' in the cmd window).
8) This data can now be used to accurately locate and modify variable regions in the protein of interest.
https://www.dropbox.com/s/bai70ev5oqksj98/Protein%20Calculator.zip
 "
"