Results
Over the summer Team Magneto worked hard and some of the preset goals were successfully accomplished. Here we show and explain you our main results.
Quick Overview
MamC-eGFP: the functionality of the eGFP part of the fusion was proven, regarding the MamC part only the expected size of the protein could be shown
MamC alone couldn’t be further characterized
eGFP: the functionality and autenticity of the protein could be shown
eFbFP: the submitted BioBrick couldn’t be successfully characterized
MamC-eGFP
MamC (BBa_K1094001) from Magnetospirillum magnetotacticum (MS-1) was tagged with enhanced green fluorescent protein (eGFP, BBa_K1094400). Between the parts a glycine linker (10 glycine residues) is added. The gene product can be used to detect localization of MamC in MS-1. The two parts were cloned together using classical cloning into pJAM1786 in E. coli. Colony PCR and gel electrophoresis was performed to ensure the insert was in the vector (Fig.1).
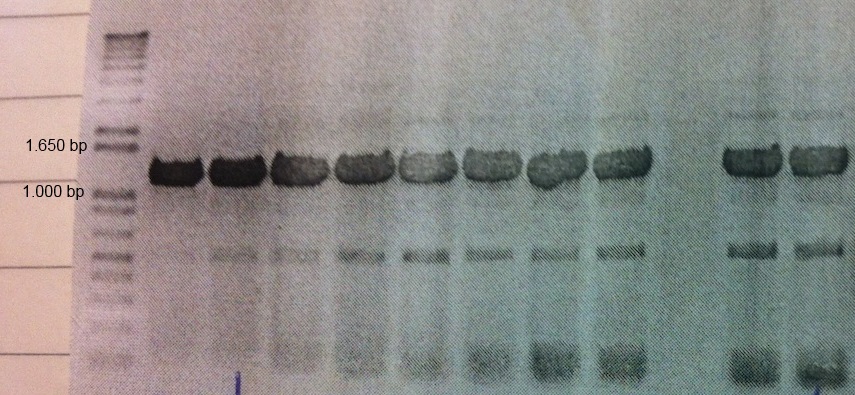
Figure 1: Colony PCR and gel electrophoresis was performed to ensure the insert was in the vector. The figure shows clear bands at around the expected size of 1.2 kb
In order to demonstrate that the fusion protein was both expressed and functional, we made an SDS page followed by western blot analysis and we also observed a fluorescence signal by using confocal microscopy. Moreover, fluorescence measurements were made on the ELISA reader.

Figure 2: Western Blot analysis of transformed E.coli cultures with anti-GFP antibody. Shows expected band of MamC-eGFP at around 45 kDa and a band for cleaved off eGFP at around 32 kDa
 "
"


