Team:USTC-Software
From 2013.igem.org
| Line 57: | Line 57: | ||
| - | <div id="video" align="center"> | + | <!--div id="video" align="center"> |
<!--embed wmode="transparent" src="https://youtube.googleapis.com/v/kKy9ILR34RA" | <!--embed wmode="transparent" src="https://youtube.googleapis.com/v/kKy9ILR34RA" | ||
type="application/x-shockwave-flash" width="100%" height="238px" style="margin-top:15px;" | type="application/x-shockwave-flash" width="100%" height="238px" style="margin-top:15px;" | ||
| Line 69: | Line 69: | ||
<p>Synthetic biology has been working on transforming target organisms, which usually means integrating new genes with an available network to achieve a high expression level of certain compounds. Nevertheless, the new-integrated genes are always not the original parts of the target metabolic network, so it is hard to predict how the new genes will affect the network. </p> | <p>Synthetic biology has been working on transforming target organisms, which usually means integrating new genes with an available network to achieve a high expression level of certain compounds. Nevertheless, the new-integrated genes are always not the original parts of the target metabolic network, so it is hard to predict how the new genes will affect the network. </p> | ||
<p>Our application aims to simulate genetic networks. The application analyzes the stability of genetic networks after introduction of exogenous genes. Meanwhile, given the original network and specific purposes, the application traces the regulative process back and gives possible regulative patterns.</p> | <p>Our application aims to simulate genetic networks. The application analyzes the stability of genetic networks after introduction of exogenous genes. Meanwhile, given the original network and specific purposes, the application traces the regulative process back and gives possible regulative patterns.</p> | ||
| - | </div> | + | </div--> |
<div id="contain"> | <div id="contain"> | ||
Revision as of 04:45, 22 August 2013
Welcome to our wiki!
Description
Synthetic biology has been working on transforming target organisms, which usually means integrating new genes with an available network to achieve a high expression level of certain compounds. Nevertheless, the new-integrated genes are always not the original parts of the target metabolic network, so it is hard to predict how the new genes will affect the network.
Our application aims to simulate genetic networks. The application analyzes the stability of genetic networks after introduction of exogenous genes. Meanwhile, given the original network and specific purposes, the application traces the regulative process back and gives possible regulative patterns.
Abstract
Map
Map is a simple and easy software to visualize your experimental data. Click to import the data and you will get exactly what you want.
Genetic Regulatory Networks
Genetic Regulatory Network(GRN) has been a major subject in recent researches of synthetic biology, and the modulation of a GRN gives rise to a variety of exciting works among iGEM programs as well as softwares assisting synthetic biology researches. Traditionally, researches of GRNs have been focused on either connecting GRNs with real parts in the registry or with experimental data. Therefore, we want to completely connect these three factors.
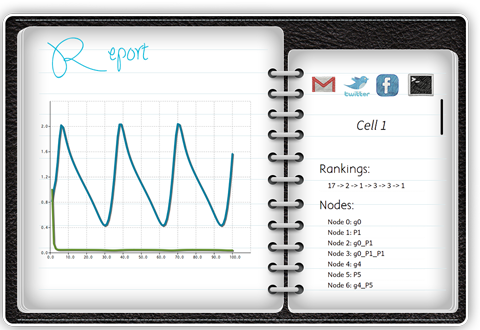
Report
Report organizes all output information in folders. You can review the simulation result while looking at the behaviors of certain genes or proteins. In addition, Report creates a web page where you can review the results on the go.
Reverse Engineering
Reverse Engineering is the process of discovering the inner technological or scientific principles of a device, object or system. Synthetic biology is a combination of biology and engineering, and with the engineering process gets more and more complicated, often it is impossible to easily understand the inside, which is the biological part of the system. Reverse engineering serves perfectly for the purpose to bring back the biological essence.

Machine Learning
Machine learning is a branch of artificial intelligence that is widely used in many disciplines such as software engineering, computer vision, finance, Neuroscience and bioinformatics. During reverse engineering, we use many machine learning techniques and algorithms to help optimize the process so that the process is faster and more accurate.
All-in-one Software
In order to fully combine biological networks, experimental data and mathematical models, we build a suite of applications that serves to solve different tasks and make them work seamlessly together. With the all-in-one software suite, you can display experimental data, extract mathematical models, understand the genetic regulatory networks and get a well-designed report of the results. We designed the user interfaces to be fun to interact with and easy to use.
Console
Console is where you manage heavy computing and complex tasks. With different buttons controlling each parameter, you can optimize the behaviors of the software. We applied evolution algorithm and machine learning techniques in the network inferences to provide the best simulation of your data.
Sand Box
SandBox displays the Genetic Regulatory Networks in a clean and interactive way, with clear connection and 3-D interaction, you will get better understanding of how genes and proteins regulating each other.
 "
"













