Team:UT Dallas/Parts
|
Experiments A) Promoter Evaluation We began by testing the relative strength of the promoters we added to the registry: pFru (BBa_K1214007) and pScr (BBa_K1214006). Our promoters and the medium constitutive promoter (BBa_K823014) were ligated with RBS-GFP, which served as our reporter molecule, and subsequently transformed into our chassis organism. The experiment we ran used glycerol stocks from this transformation, inoculated in 5mL of chloramphenicol broth overnight. The resulting cell cultures were then used for Flow cytometry analysis and microscope imaging. 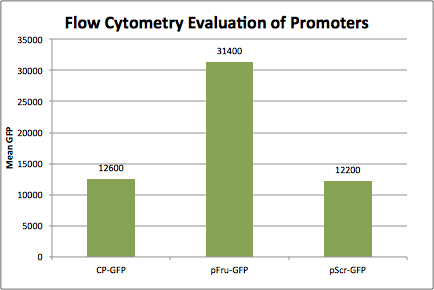 Background Cp-GFP (left) versus GFP Expression at 395nm (right) 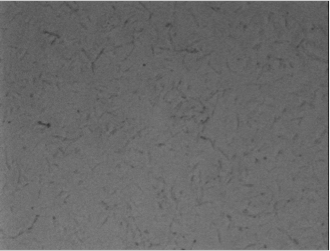 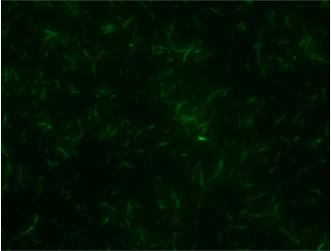 Background pScr-GFP (left) versus GFP Expression at 395nm (right)  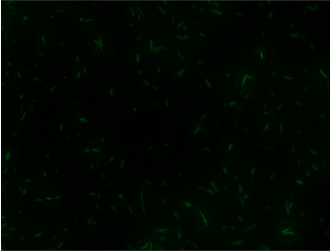 Background pFru-GFP (left) versus GFP Expression at 395nm (right) 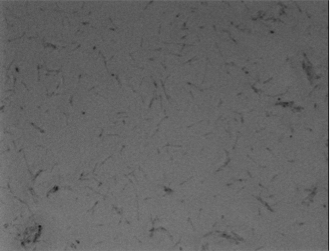 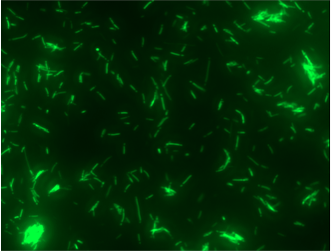
Our team submitted two new repressors to the registry that work in conjunction with the aforementioned promoters. These repressors are Cra (BBa_K1214008) and ScrR (BBa_K1214005). In theory, these should be inducible repressors that repress their respective promoters. Cra (also known as FruR) will repress pFru until Fructose is introduced, wherein the repressor will unblock pFru and promotion will continue. The same relationship is expected between ScrR and pScr except that promotion is induced by sucrose in this case. These repressors were transformed to make four systems, with the medium constitutive promoter indicated by Cp:1. pFru-RBS-GFP-Cp-RBS-ScrR 2. pFru-RBS-GFP-Cp-RBS-Cra 3. pScr-RBS-GFP-Cp-RBS-ScrR 4. pScr-RBS-GFP-Cp-RBS-Cra These were plated, incubated overnight. Seeing as the plates were from a transformation, not all colonies are guaranteed to contain the completed system. Thus, the GFP expression on these plates would be less than the true GFP expression of the system and they were not used in our characterization.. Colonies were then picked from these plates and inoculated in 5mL of chloramphenicol broth overnight. This inoculation was used to perform Flow cytometry analysis of the repressors. It should be noted that the promotion of the repressors is dictated by Cp more than pFru or pScr, and in conjunction with the aforementioned promoter data it can be seen that pFru may cause greater downstream promotion than Cp but was not used due to conflicts with repression. However, the data obtained shows the expected repression patterns, especially in relation to the specificity of the repressors. [[File:FlowpScrrepression.png]] [[File:FlowpFrurepression.png]] These repressors are shown to greatly reduce the GFP seen in the systems without repressors. pFru expression of GFP under the repressor was reduced at least five-fold while pScr expression of GFP was reduced at least ten-fold. There is also evidence of the repressor’s specificity shown in the difference between the promoter’s GFP expression depending on whether Cra or ScrR was in the system. The sucrose promoter was repressed approximately four times as much by ScR than by Cra. pFru was repressed roughly 20% more by Cra than by ScrR. This data supports the preexisting claim that the repressors are more specific to their particular promoters. This is important to consider when using our biobricks in a system. Preliminary tests on the inducibility of the repressors was performed but the data was inconclusive. |
 "
"