Team:Imperial College/Collaboration
From 2013.igem.org
Contents |
Collaborations
If you would like to collaborate just contact us via Twitter @imperialigem
Helpers at SB6
The BioBricks Foundation Synthetic Biology Conference Series (SBx.0 Conference) is the world’s foremost professional meeting in the field of synthetic biology. The BioBricks Foundation is the proud organizer of the conference series and this year Imperial had the honour of hosting the conference. SB6 ran 9th-11th July 2013. We were helpers at the conference, working behind the scenes to help delegates and in collaboration with the event organisers we ensured everything ran smoothly.
We were also lucky enough to see some great talks, and enjoyed networking amongst the synbio community! The buzz of SB6.0 gave us a boost and the inspiration for our iGEM project.
Presented at the YSB1.0 meeting
We presented at the very first Young Synthetic Biologists meeting at the Wellcome Trust. YSB1.0 was designed as an extension of the UK iGEM team meetup that has been organised in previous years. We had a great time and loved sharing our ideas, networking and having fun with the rest of the UK iGEM community. As an additional bonus, Randy Rettberg, one of the co-founders and current president of the iGEM foundation gave us an inspirational talk.
Saint Paul's School iGEM team
We met the team from St.Paul's School when we went to the Young Synthetic Biology event. We were impressed by their project of the previous year and got chatting to them. As school students they were keen to see some University Labs and discuss their project further so we invited them for a tour of the facilities at Imperial. We hope the tour was enjoyable and that the tips we gave them come in useful in their entry this year.
Yale: PLA-degradation
As soon as we found out Yale iGEM team was synthesizing poly lactic acid(PLA) this year, we contacted them and sent them our PLA degradation parts to test on their product. We did this to further characterise the part and see if Biobricks behave the same on both sides of the Atlantic! When Yale optimises PLA production, they will send some of their product to break down. Breaking down PLA is a second biological bioplastic recycling system we are working on, capable of producing the monomers required to chemically resynthesise it.
UCL:Beta-Amyloid Degradation
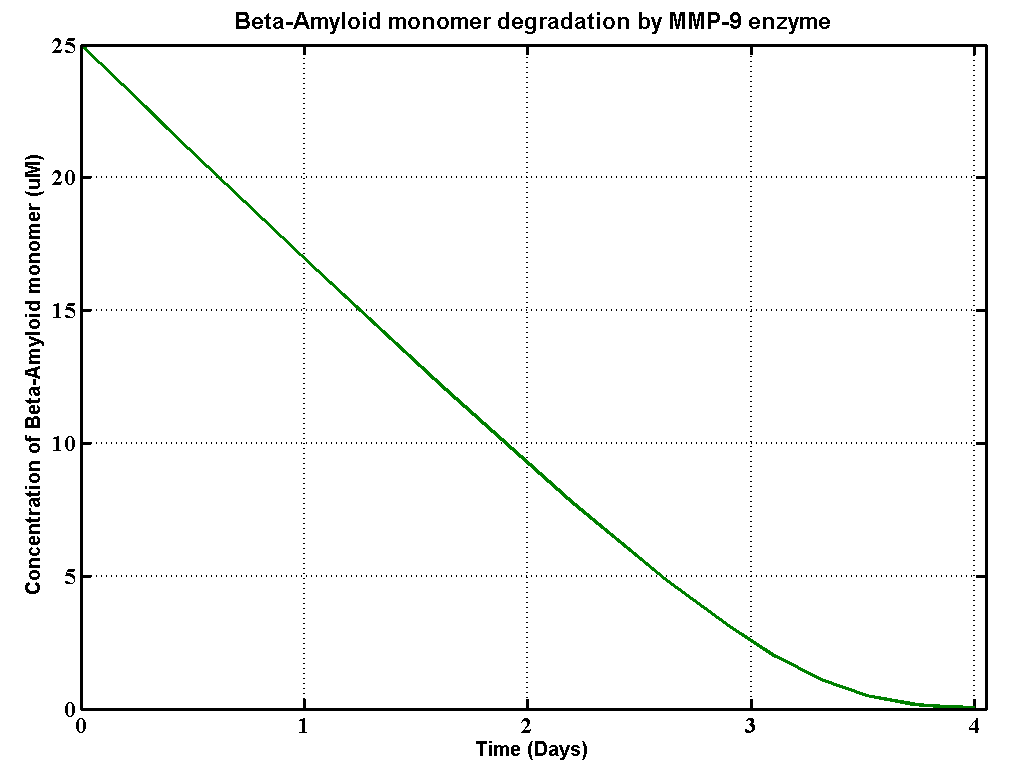
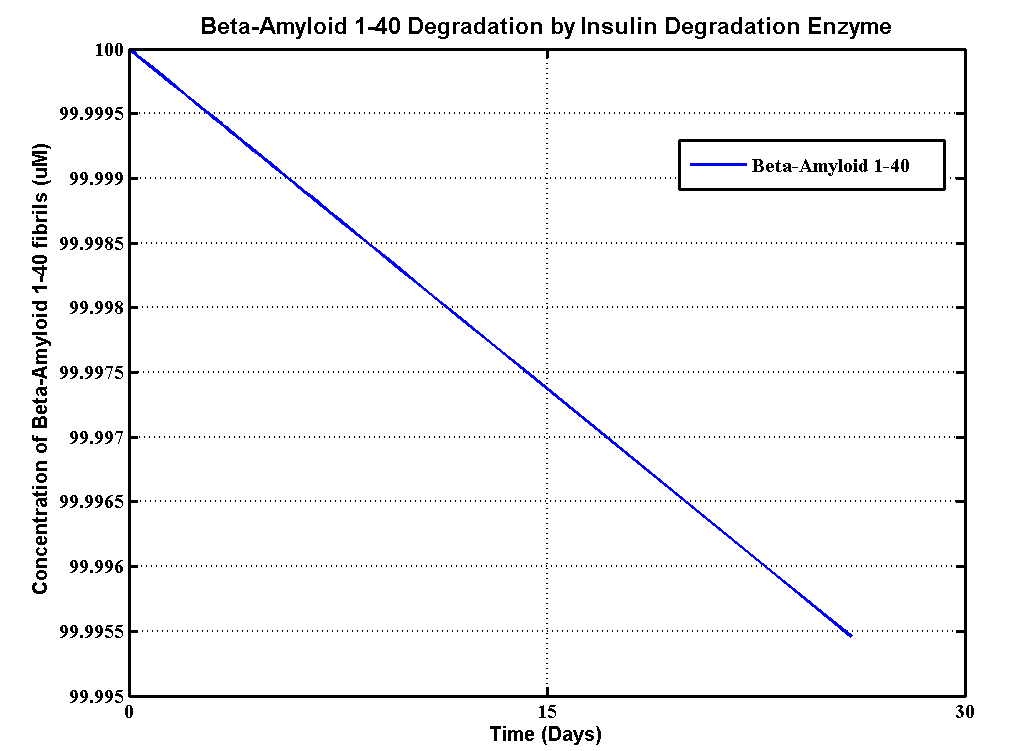
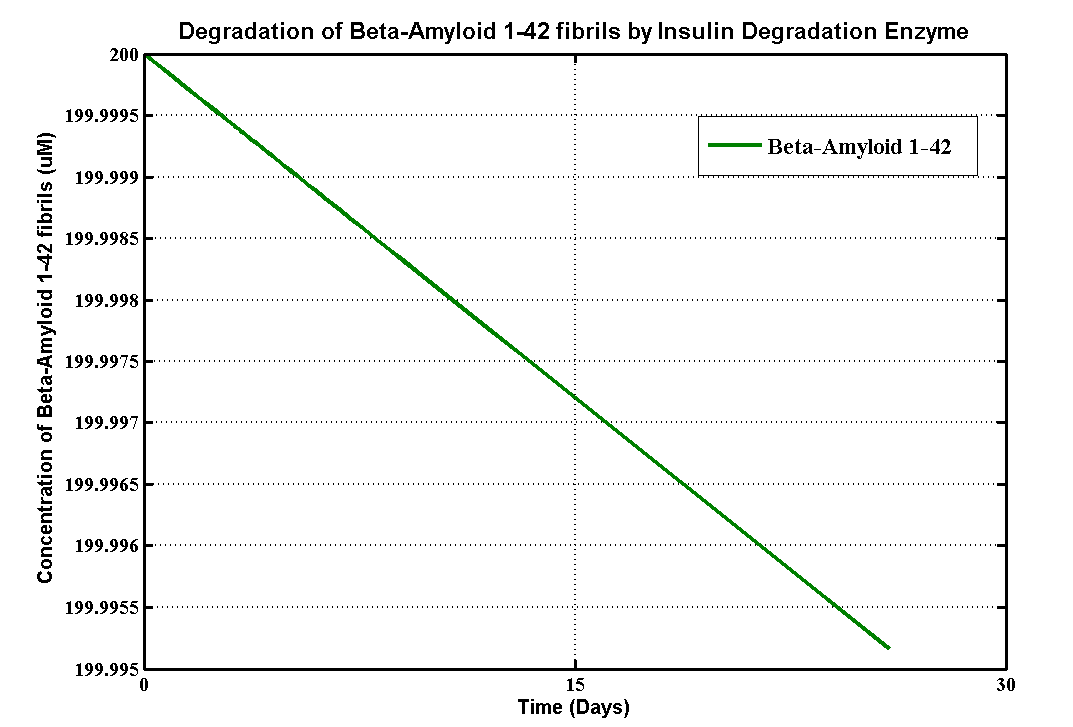
Apart from sending beta-amyloid degradation part Proteinase K to UCL,we also built a degradation model to help UCL 2013 iGEM team with their beta-amyloid degradation assays. Beta-Amyloid is known as the component of Amyloids Plaque in association with Alzheimer's disease (AD), which UCL iGEM is targetting this year. This model, based MATLAB extension Simbiology, provides information about how efficient MMP-9 enzyme can degrade beta-amyloid plaques, so that they use the simulation results to modify their amyloid plaque degradation assays.
Degradation Assays of UCL 2013 iGEM team
The details of the assays are here. There are three different substrates which are soluble aggregated β-amyloid (Aβ),aggregated β-amyloid (Aβ) fibrils 1-40 and aggregated β-amyloid (Aβ) fibrils 1-42. Here is the table that defines the initial concentrations of all substrates and the enzyme from the assays.
| Species | Concentration | Units |
|---|---|---|
| soluble Aβ | 25 | uM |
| Aβ fibrils 1-40 | 200 | uM |
| Aβ fibrils 1-42 | 100 | uM |
| MMP-9 | 0.2 | uM |
Methods
The Model
Explanation
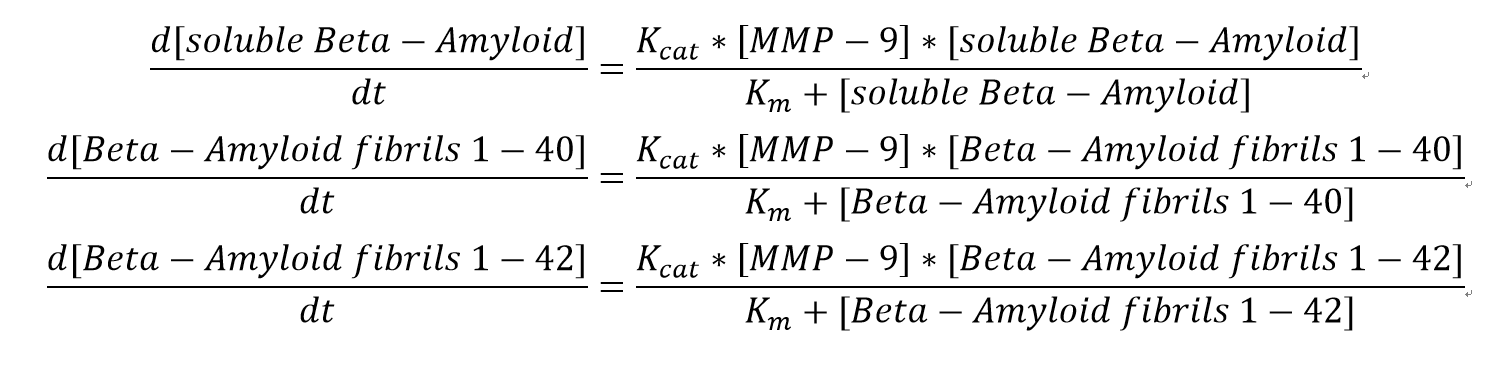
Equations
We choose simple Michalis-Menten mechanism for Beta-Amyloid degradation. The ODEs for all substrate are:
The Km values here are for each substrate correspondingly whereas Kcat values are for MMP-9 in each assay.
Assumption
Although Matrix metallopeptidase 9 (MMP-9) is a well characterised enzyme, there is no clear kinetic data for degrading soluble Beta-Amyloids and its fibrils.
Results
References
NRP-UEA_Norwich
We sent NRP-UEA Norwich iGEM team soil samples collected from London, in order for them to identify antimycin-producing bacteria inhabiting in the samples.
| iGEMHS Team: Saint Paul's | ||
|---|---|---|
| Presented at the Young Synthetic Biologists meeting | ||
| We were helpers at SB6 | ||
| We transformed and sent for sequencing BioBrick parts BBa_C0061 and BBa_C0062 from our 2013 parts distribution. We were prepared to send the physical DNA to the team, however this became unnecessary since following our transformation advice the team was successful in obtaining the DNA from their own distribution. | ||
| We contributed comments and suggestions to the teams characterisation survey http://www.surveymonkey.com/s/M6SVLZ6 | ||
| We responded to the NRP_UEA-Norwich iGEM team request for soil samples. | ||
| Gave B. subtilis promoter advice to the Chicago iGEM team.
|
 "
"