Team:Newcastle/Parts/l form switch
From 2013.igem.org

Rod to L-form Switch BioBrick
Purpose and Justification
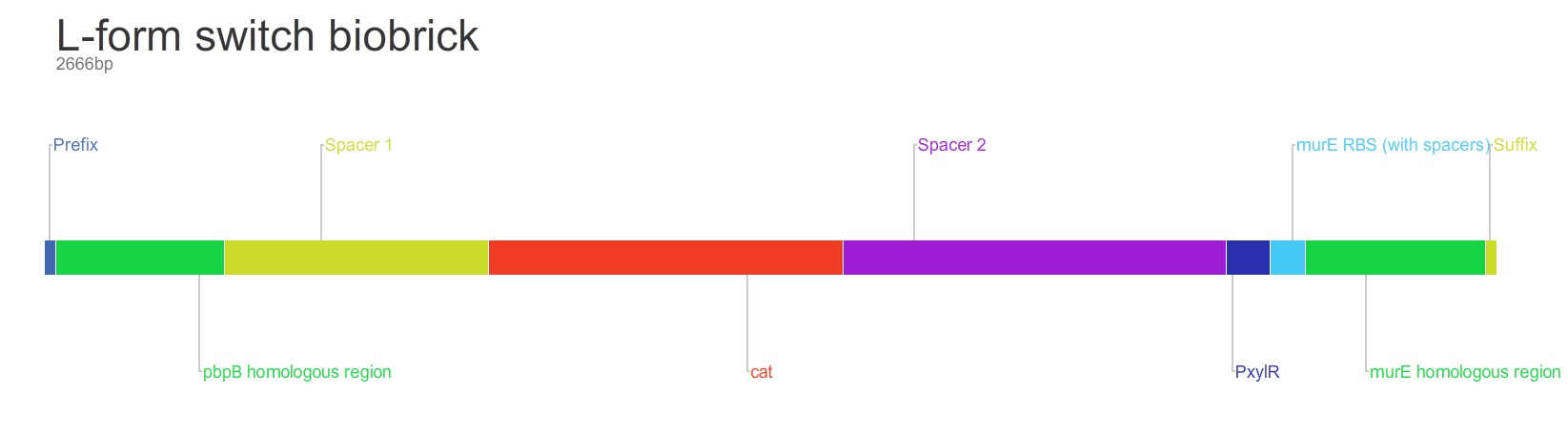
The purpose of this main ‘keystone’ biobrick is to facilitate the switching of Bacillus subtilis cells from rod-shape to L-form, wall-less cells. Furthermore, this ‘switch’ enables L-form cells to return to rod-shape when required. This is facilitated through the introduction of a xylose-controlled promoter (PxylR) upstream of the murE gene, which is involved in the biosynthesis of peptidoglycan. The presence, or absence of a cell wall can be controlled through the presence, or absence of xylose, respectively. The antibiotic resistance marker chloramphenicol acetyl-transferase (cat) is upstream of this promoter region to allow for the selection of cells in which this system is integrated.
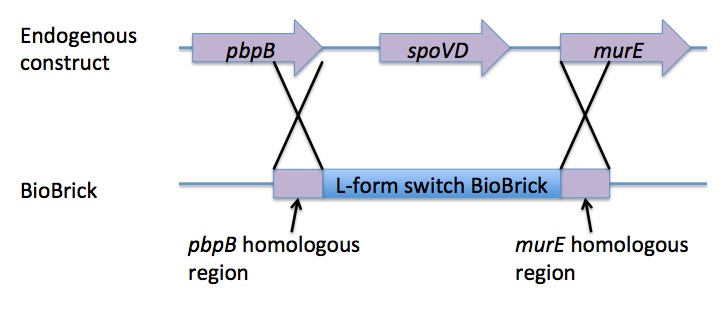
Homologous recombination allows this BioBrick to be integrated into the chromosome of B. subtilis. The cat and PxylR sequences are flanked by sequences that are homologous with regions of the B. subtilis chromosome, which allows for insertion of the BioBrick at the intended loci. ~300bp at the 5’ end of the BioBrick is homologous with the end of the pbpB gene. Similarly ~300bp at the 3’ end of the biobrick is homologous with the start of the murE gene.
Integration of the BioBrick facilitates xylose-mediated control over the expression of murE. This cascades through biosynthetic pathways to enable control over peptidoglycan synthesis and thus control over cell wall production. Simply, in B. subtilis cells that have integrated the BioBrick into their chromosome, only in the presence of xylose is the cell wall produced. When xylose is not present cells lose their cell wall and survivors adopt the L-form phenotype.
The L-form BioBrick BBa_K1185000 can be found on the Registry of Standard Biological Parts.

Design
The L-form switch BioBrick is based upon the regulatory sequence controlling expression of murE in B. subtilis LR2 - a strain capable of L-form state. In this B. subtilis strain the peptidoglycan synthesis pathway required for the cell wall can be disrupted through control of murE expression - responsible for the synthesis of enzymes involved in peptidoglycan precursor biosynthesis. A xylose-contolled PxylR promoter upstream of murE facilitates this control. Upstream of this promoter the LR2 strain also contains a cat gene - conferring chloramphenicol resistance. These genetic components differentiate the B. subtilis LR2 chromosome from that of B. subtilis 168. On the LR2 chromosome these genetic components take the place of the spoVD gene (involved in sporulation) in B. subtilis 168. It is these genetic elements that have been utilised to create a BioBrick that facilitates the switching of B. subtilis cells from rod state to L-form, and back again. The regions flanking the cat gene and the PxylR promoter - part of the pbpB coding sequence at the 5' end of this region of DNA and part of the murE coding sequence at the 3' end - are also of vital importance, allowing for integration of the BioBrick into the chromosome of B. subtilis via homologous recombination.
In order to package the genetic elements within the B. subtilis chromosome that enable this strain to transition from rod to L-form, the full sequence of the region between the pbpB and the murE genes had to be identified. The sequence for the L-form switch region was obtained through a de novo assembly (using Sequencher 5.1 DNA sequencing software) of sequence data gained from sequencing between the pbpB and murE genes on the B. subtilis LR2 chromosome (using designed primers) with reads produced by high throughput sequencing of the full B. subtilis LR2 genome. In particular, reads that
 "
"