Team:SCU China/Modeling
From 2013.igem.org
Zhangbingbio (Talk | contribs) |
Zhangbingbio (Talk | contribs) |
||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{SCU-head}} | {{SCU-head}} | ||
| - | {| style="text-align: | + | <html> |
| + | <body> | ||
| + | <div id="SCU"> | ||
| + | <ul style="font-family:Verdana;;"> | ||
| + | <!-- CSS Tabs --> | ||
| + | <li><a href="https://2013.igem.org/Team:SCU_China"><span style="width:90px; text-align: center;font-size: small;">Home</span></a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:SCU_China/Team"><span style="width:90px;text-align: center;font-size: small;">Team</span></a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:SCU_China/Project"><span style="width:90px;text-align: center;font-size: small;">Project</span></a></li> | ||
| + | <li><a class="this" href="https://2013.igem.org/Team:SCU_China/Modeling"><span style="width:100px;text-align: center;font-size: small;">Modeling</span></a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:SCU_China/Parts"><span style="width:100px;text-align: center;font-size: small;">Parts</span></a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:SCU_China/Safety"><span style="width:100px;text-align: center;font-size: small;">Safety</span></a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:SCU_China/Notebook"><span style="width:100px;text-align: center;font-size: small;">Notebook</span></a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:SCU_China/Attributions"><span style="width:100px;text-align: center;font-size: small;">Attributions</span></a></li> | ||
| + | <li><a href="https://2013.igem.org/Main_Page"><span style="width:100px;text-align: center;font-size: small;">iGEM 2013</span></a></li> | ||
| + | |||
| + | |||
| + | |||
| + | </ul> | ||
| + | </div> | ||
| + | |||
| + | </body></html> | ||
| + | |||
| + | | ||
| + | |||
| + | {|style="text-align:justify; text-justify:inter-ideograph; width: 800px" border="0" cellspacing="1" cellpadding="20" align="center" | ||
|- | |- | ||
| | | | ||
| + | '''1 Introduction''' | ||
| + | |||
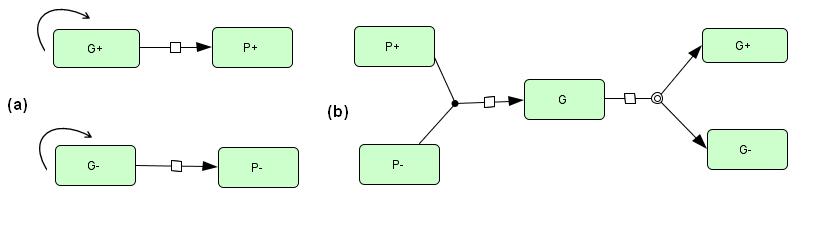
| + | <div style="float:left">In our project, we intended to construct two groups of differentiated ''E. coli''. One imitated the male multicellular organism, while the other was for the female (called G+ cells and G- cells respectively). In this project, the modified G+ and G- cells would be cultivated separately first, and then put together. Therefore, we had to simulate the two phases. The first part focused on gamete forming (from G+/G- to P+/P-), while the second part was forming of the “zygotes” (G cells, from P+ and P- cells) and the separation of them to G+ and G-. These processes are shown in figure 1. | ||
| + | [[file:SCU-m-1.jpg]] | ||
| + | Fig 1 Diagrams of 3 cultivating system. (a) G<sub>+</sub> and G<sub>- </sub>cultivated separately. (b) The mixed system. | ||
| + | |||
| + | We built two models to describe and simulate our experiment: | ||
| + | |||
| + | '''Model of transition into gametes:''' We described the transition process of ''E. coli ''and then simulate the increase and decrease of ''cre'' recombinase. | ||
| + | |||
| + | '''Model of combination of gametes: '''We used Markov process to describe the combination of two gametes, which started with circumstance 1 (two individual cells) and ended up with circumstance 2 (conjugation between two gametes get result of one zygote). | ||
| + | |||
| + | '''2 Transition into gametes.''' | ||
| + | |||
| + | '''2.1 Model hypothesis''' | ||
| + | |||
| + | 1. Although signal molecule growth rate of each ''E. coli'' changes, considering the number of colonies of ''E. coli'', we ignore the growth rate changes. | ||
| + | |||
| + | 2. For a quorum sensing system, ''cre'' gene was activated to adapt the changes of the environment only when the amount of signal molecule reached a certain threshold. So we could assume that the concentration in each ''E. coli'' is nearly the same. | ||
| + | |||
| + | '''2.2 Model''' | ||
| + | |||
| + | 2.2.1 The amount of ''cre'' before transition. When the amount of ''cre'' reaches a certain number, the transition occurs. So finding the probabilities for amounts of ''cre'' is essential. | ||
| + | [[file:SCU-m-2.GIF]] | ||
| + | We let N be the amount of E. ''coli''. N<sub>max</sub> is a coefficient constant which refers the maximum amount that the environment can accommodate. [[file:SCU-m-21.GIF]] is a growth constant. | ||
| + | |||
| + | <div style="float:left">According to the previous assumption, the amount of signal molecule can be expressed by N<sub>sign.</sub>[[file:SCU-m-3.GIF]]Where K<sub>sig</sub> is a coefficient constant that represents the amount of the signal molecule produced by one ''E. coli''. We know from the experimental data that when OD600 reaches 0.5, | ||
| + | [[file:SCU-m-4.GIF]] | ||
| + | So we make k<sub>sig</sub> =4.82×10<sup>8</sup> | ||
| + | When the amount of signal molecule reaches the threshold value, the ''cre'' gene is activated to start generating the ''cre''. We let t<sub>1</sub> be the point when the amount of signal molecule reaches the threshold value. We let t<sub>2</sub> be the point when the amount of ''cre'' reaches a certain number and the | ||
| + | |||
| + | ''E. coli'' starts out to transform. So here is the equation to describe the ''cre.'' | ||
| + | |||
| + | [[file:SCU-m-5.GIF]] | ||
| + | |||
| + | T is the half-life. K<sub>c</sub> is a growth constant. | ||
| + | |||
| + | If we let k<sub>c</sub>=6.1538×10<sup>-11</sup>, t<sub>2</sub>=1400, then we get figure 2, 3, 4. | ||
| + | |||
| + | [[file:SCU-m-6.GIF|500px]] | ||
| + | |||
| + | Fig 2 Amount of ''E. coli'' (k<sub>c</sub>=6.1538×10<sup>-11</sup>,t<sub>2</sub>=1400) | ||
| + | |||
| + | [[file:SCU-m-7.GIF|500px]] | ||
| + | |||
| + | Fig 3 Amount of signal molecule (k<sub>c</sub>=6.1538×10<sup>-11</sup>,t<sub>2</sub>=1400) | ||
| + | |||
| + | [[file:SCU-m-8.GIF|500px]] | ||
| + | |||
| + | Fig 4 Amount of ''cre'' (k<sub>c</sub>=6.1538×10<sup>-11</sup>,t<sub>2</sub>=1400) | ||
| + | |||
| + | If we let k<sub>c</sub> =6.1538×10<sup>-11</sup>,t<sub>2</sub>=300, then we get figure 5, 6, 7. | ||
| + | |||
| + | [[file:SCU-m-9.GIF|500px]] | ||
| + | |||
| + | Fig 5 Amount of ''E. coli'''' '''''( k<sub>c</sub> =6.1538×10<sup>-11</sup>,t<sub>2</sub> | ||
| + | |||
| + | [[file:SCU-m-10.GIF|500px]] | ||
| + | |||
| + | Fig 6 Amount of signal molecule (k<sub>c</sub> =6.1538×10<sup>-11</sup>,t<sub>2</sub>=300) | ||
| + | |||
| + | [[file:SCU-m-11.GIF|500px]] | ||
| + | |||
| + | Fig 7 Amount of ''cre'' (k<sub>c</sub> =6.1538×10<sup>-11</sup>,t<sub>2</sub>=300) | ||
| + | |||
| + | If we let k<sub>c</sub> be nine different values: 0.0615×10<sup>-8</sup>, 0.1231×10<sup>-8</sup> , 0.1846×10<sup>-8</sup>, 0.2462×10<sup>-8</sup>, 0.3077×10<sup>-8</sup>, 0.3692×10<sup>-8</sup>, 0.4308×10<sup>-8</sup>, 0.4923×10<sup>-8</sup>, 0.5538×10<sup>-8</sup>, then we get figure 8, | ||
| + | |||
| + | [[file:SCU-m-13.GIF|500px]] | ||
| + | |||
| + | Fig 8 Amount of ''cre''(t<sub>2</sub>=1400) | ||
| + | |||
| + | According to this figure, although we do not know the exact value of kc, we are sure that, despite of the value of k<sub>c</sub>, the increasing speed of ''cre'' will be sharply accelerated in 600 minutes and will be slow down to zero in 1000 minutes. These figures implicate that we could test the fluorescence after about 600- minute culture. | ||
| + | |||
| + | 2.2.2 The probability of transition | ||
| + | |||
| + | Then we estimate that when the concentration of enzymes is x, the probability of transition is the thing that determine the cause of G+/G- transferring to P+/P-. Using the previous results, we know that concentrations of ''cre'' at each time. | ||
| + | |||
| + | Based on the assumption 2, we know that concentrations of ''cre'' for the each ''E. coli ''(ξ<sub>1</sub>,…, ξ<sub>n</sub>…) are random, independent and identically distributed. | ||
| + | |||
| + | Drawn N samples from the data obtained in our experiments, and then: | ||
| + | |||
| + | Step 1. Calculating the sample's expectations E (ξ) =μ, variance D (ξ) =σ<sup>2 </sup>and standardization [[file:SCU-m-14.GIF]] | ||
| + | |||
| + | Step 2. By the Lindbergh - Levi central limit theorem,[[file:SCU-m-15.GIF]] we get function of concentration: [[file:SCU-m-16.GIF]] | ||
| + | |||
| + | So we can get the concentration [[file:SCU-m-17.GIF]] after standardized variables, and probability of unturned bacterial under this concentration is Φ(x). Then the ratio of transformation is 1-Φ(x). | ||
| + | |||
| + | Since our test has not been completed, the lacking of this part of the data led us not to reach concrete results. | ||
| + | |||
| + | '''3 Mix two gametes group''' | ||
| + | |||
| + | '''3.1 Model hypothesis''' | ||
| + | |||
| + | 1) The broth and culture condition for each gamete and zygote is the same. | ||
| + | |||
| + | 2) The nutrition of each plate for bacteria growth is sufficient and there is no stress from environment. | ||
| + | |||
| + | 3) It takes some time for zygote to differentiate into gametes of different gender, we assume that this process lasts for a long time and the differentiation of zygote is after the combination of gametes. | ||
| + | |||
| + | '''3.2 modeling''' | ||
| + | |||
| + | 3.2.1Gametes combination | ||
| + | |||
| + | After G+ and G- cells has been cultured separately and converted into P+ and P- , two culture systems are mixed to form a new system. Five different kinds of cells as mentioned above exist in the system, and only the G+ and G- cells have the ability to proliferate while the “zygotes”, G cells can also divide. The moving traces of G cells in the petri dish are independent of each other. Because the medium in the petri dish is homogeneous, its trajectory can be approximately viewed as Brownian motion. | ||
| + | |||
| + | Simply we can take the whole petri dish as a region "E" which has a circle boundary. The collection of locations which P+(or P-)moving through all the place called state space(That is the entire dish area). Since Brownian motion is essentially a Markov random process, its Markov property: | ||
| + | | ||
| + | <div style="float:left">For any | ||
| + | [[file:SCU-m-18.GIF]] | ||
| + | we have [[file:SCU-m-19.GIF]] let locations of P+ and P- were n1 and n2, then P+ and P- will combine when p[x(n1)]=p[x(n2)] at the same time.</div> | ||
| + | | ||
| + | So Markov process is describing a probability of that gametes reach each state (position). In this process, two gametes become zygote through collision. Therefore when at the same time t, P+ and P- reach the same state E0 (That collide with each other form a zygote). Thus, we describe a stochastic process with complete binding process. We can calculate the gametes P+ and P- where at the states of e1 and e2 in initial moment by a Markov probability equation: | ||
| + | |||
| + | 1) How long it takes for gametes to combine into a zygote? | ||
| + | |||
| + | 2) The combination probability at time t. | ||
| + | |||
| + | 3.2.2 Zygote division | ||
| + | When the entire population of gamete combine into zygotes after a period of culture, these zygotes will redivide into G+ and G- cells. This is a natural process of cell division. | ||
|} | |} | ||
| + | | ||
Latest revision as of 20:05, 27 September 2013

|
1 Introduction In our project, we intended to construct two groups of differentiated E. coli. One imitated the male multicellular organism, while the other was for the female (called G+ cells and G- cells respectively). In this project, the modified G+ and G- cells would be cultivated separately first, and then put together. Therefore, we had to simulate the two phases. The first part focused on gamete forming (from G+/G- to P+/P-), while the second part was forming of the “zygotes” (G cells, from P+ and P- cells) and the separation of them to G+ and G-. These processes are shown in figure 1.
We built two models to describe and simulate our experiment: Model of transition into gametes: We described the transition process of E. coli and then simulate the increase and decrease of cre recombinase. Model of combination of gametes: We used Markov process to describe the combination of two gametes, which started with circumstance 1 (two individual cells) and ended up with circumstance 2 (conjugation between two gametes get result of one zygote). 2 Transition into gametes. 2.1 Model hypothesis 1. Although signal molecule growth rate of each E. coli changes, considering the number of colonies of E. coli, we ignore the growth rate changes. 2. For a quorum sensing system, cre gene was activated to adapt the changes of the environment only when the amount of signal molecule reached a certain threshold. So we could assume that the concentration in each E. coli is nearly the same. 2.2 Model 2.2.1 The amount of cre before transition. When the amount of cre reaches a certain number, the transition occurs. So finding the probabilities for amounts of cre is essential.
File:SCU-m-2.GIF
We let N be the amount of E. coli. Nmax is a coefficient constant which refers the maximum amount that the environment can accommodate. According to the previous assumption, the amount of signal molecule can be expressed by Nsign.File:SCU-m-3.GIFWhere Ksig is a coefficient constant that represents the amount of the signal molecule produced by one E. coli. We know from the experimental data that when OD600 reaches 0.5,
File:SCU-m-4.GIF So we make ksig =4.82×108 When the amount of signal molecule reaches the threshold value, the cre gene is activated to start generating the cre. We let t1 be the point when the amount of signal molecule reaches the threshold value. We let t2 be the point when the amount of cre reaches a certain number and the E. coli starts out to transform. So here is the equation to describe the cre. T is the half-life. Kc is a growth constant. If we let kc=6.1538×10-11, t2=1400, then we get figure 2, 3, 4. Fig 2 Amount of E. coli (kc=6.1538×10-11,t2=1400) Fig 3 Amount of signal molecule (kc=6.1538×10-11,t2=1400) Fig 4 Amount of cre (kc=6.1538×10-11,t2=1400) If we let kc =6.1538×10-11,t2=300, then we get figure 5, 6, 7. Fig 5 Amount of E. coli' ( kc =6.1538×10-11,t2 Fig 6 Amount of signal molecule (kc =6.1538×10-11,t2=300) Fig 7 Amount of cre (kc =6.1538×10-11,t2=300) If we let kc be nine different values: 0.0615×10-8, 0.1231×10-8 , 0.1846×10-8, 0.2462×10-8, 0.3077×10-8, 0.3692×10-8, 0.4308×10-8, 0.4923×10-8, 0.5538×10-8, then we get figure 8, Fig 8 Amount of cre(t2=1400) According to this figure, although we do not know the exact value of kc, we are sure that, despite of the value of kc, the increasing speed of cre will be sharply accelerated in 600 minutes and will be slow down to zero in 1000 minutes. These figures implicate that we could test the fluorescence after about 600- minute culture. 2.2.2 The probability of transition Then we estimate that when the concentration of enzymes is x, the probability of transition is the thing that determine the cause of G+/G- transferring to P+/P-. Using the previous results, we know that concentrations of cre at each time. Based on the assumption 2, we know that concentrations of cre for the each E. coli (ξ1,…, ξn…) are random, independent and identically distributed. Drawn N samples from the data obtained in our experiments, and then: Step 1. Calculating the sample's expectations E (ξ) =μ, variance D (ξ) =σ2 and standardization File:SCU-m-14.GIF Step 2. By the Lindbergh - Levi central limit theorem,File:SCU-m-15.GIF we get function of concentration: File:SCU-m-16.GIF So we can get the concentration File:SCU-m-17.GIF after standardized variables, and probability of unturned bacterial under this concentration is Φ(x). Then the ratio of transformation is 1-Φ(x). Since our test has not been completed, the lacking of this part of the data led us not to reach concrete results. 3 Mix two gametes group 3.1 Model hypothesis 1) The broth and culture condition for each gamete and zygote is the same. 2) The nutrition of each plate for bacteria growth is sufficient and there is no stress from environment. 3) It takes some time for zygote to differentiate into gametes of different gender, we assume that this process lasts for a long time and the differentiation of zygote is after the combination of gametes. 3.2 modeling 3.2.1Gametes combination After G+ and G- cells has been cultured separately and converted into P+ and P- , two culture systems are mixed to form a new system. Five different kinds of cells as mentioned above exist in the system, and only the G+ and G- cells have the ability to proliferate while the “zygotes”, G cells can also divide. The moving traces of G cells in the petri dish are independent of each other. Because the medium in the petri dish is homogeneous, its trajectory can be approximately viewed as Brownian motion. Simply we can take the whole petri dish as a region "E" which has a circle boundary. The collection of locations which P+(or P-)moving through all the place called state space(That is the entire dish area). Since Brownian motion is essentially a Markov random process, its Markov property: For any
we have File:SCU-m-19.GIF let locations of P+ and P- were n1 and n2, then P+ and P- will combine when p[x(n1)]=p[x(n2)] at the same time.
So Markov process is describing a probability of that gametes reach each state (position). In this process, two gametes become zygote through collision. Therefore when at the same time t, P+ and P- reach the same state E0 (That collide with each other form a zygote). Thus, we describe a stochastic process with complete binding process. We can calculate the gametes P+ and P- where at the states of e1 and e2 in initial moment by a Markov probability equation: 1) How long it takes for gametes to combine into a zygote? 2) The combination probability at time t. 3.2.2 Zygote division When the entire population of gamete combine into zygotes after a period of culture, these zygotes will redivide into G+ and G- cells. This is a natural process of cell division. |
 "
"