Team:SCU China/Project
From 2013.igem.org

Sexual differentiation is the most common and natural phenomenon in multicellutar organism, Usually, two genders’ combination through fertilization is essential for their proliferation. However, in the most primitive organisms, no sexual differentiation existed. With the evolution from protist to metazoan such as animals, definite sexual differentiation came into being. So, there is a question that when and how sexual differentiation happened. The special F factor in E. coli enables its gene communication and determination of the sex what we defined as “female” and “male”. Though this kind of gene communication is not genuinely sexual differentiation, we could make a hypothesis that the F factor is the transitory stage to true sexual differentiation. So we want to imitate sexual differentiation in unicellular organism to help us understand the process of sexual differentiation. We believe that this imitation would be very interesting and attract more people to do related research.
BackgroundAt first, we want to introduce some background in the field of using bacteria to imitate the whole developmental procedure from a single cell to a well-organized organism.
Life begins from fertilization according to developmental biology. After that, the development process includes differentiation between Germline and Soma, cell migration, pattern formation and so on. The whole course is self-organized. Imitations of above five steps except fertilization have been explored by 2007 Paris(https://2007.igem.org/Paris), 2011 USTC(https://2011.igem.org/Team:USTC-China), Nature 434,1130-34,28 April 2005 ([http://www.nature.com/nature/journal/v434/n7037/abs/nature03461.html]) and 2008 USTC(https://2008.igem.org/Team:USTC/Project), respectively. However, no comprehensive studies on the imitations of gametogenesis and sexual differentiation.
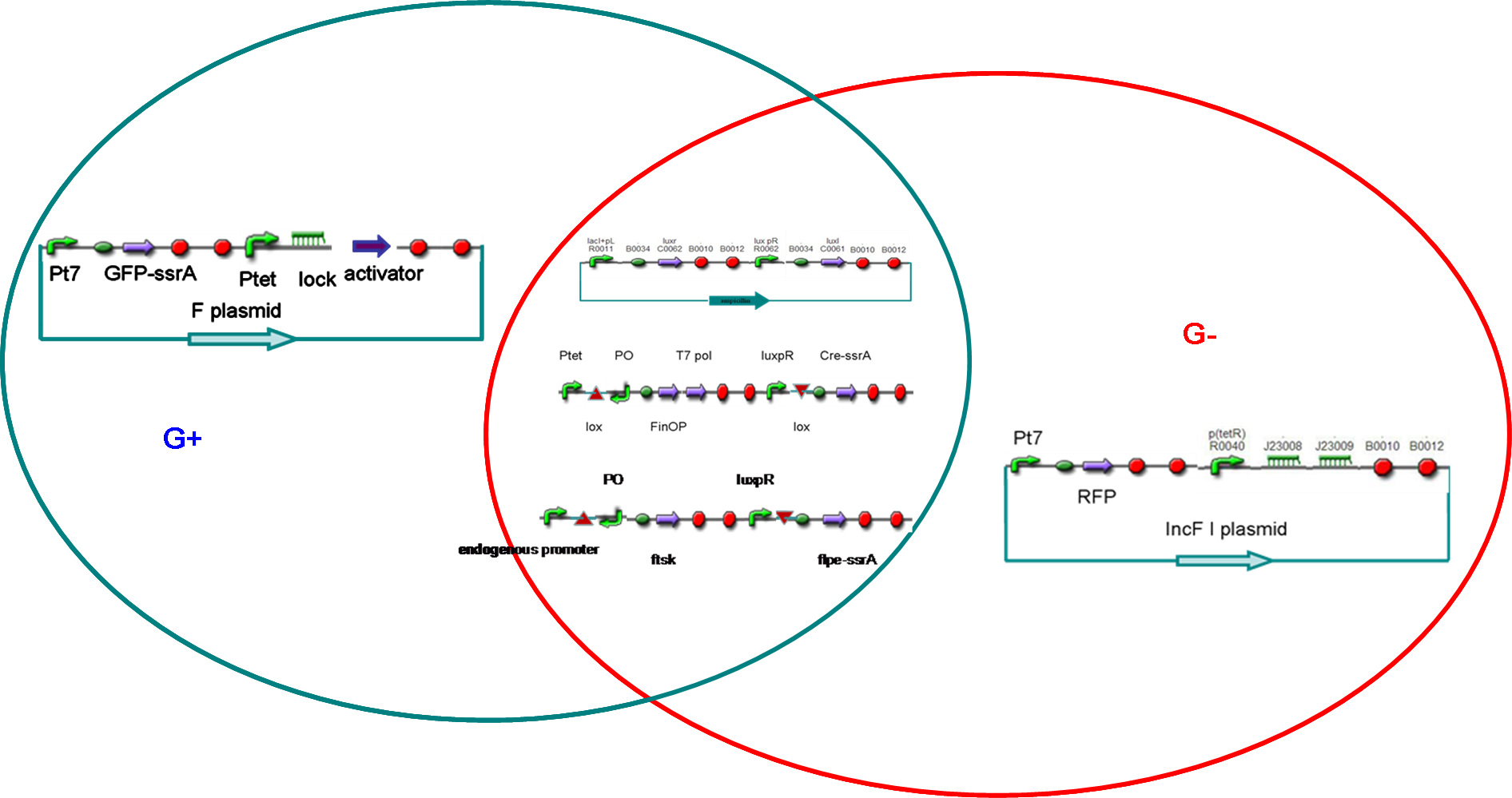
Our project designIn our project, we intended to construct male and female E. coli separately (called G+ and G- cells). According the natural characteristic of E.coli, the G+ cells possessed F plasmids. On the other hand, the G- cells were constructed by using another plasmid incompatible with F factor. When cultured separately, the G+/G- cells would divide and reach a threshold of cell density . Then, the quorum sensing system would turn on its responding promoters, and make G+/G- differentiate into gametes (P+ and P-, respectively), which could not divide any more but were capable of transferring or accepting plasmid.
After that, male and female E. coli were mixed. Just like fertilization, the male gametes would recognize the female cells and began to transfer modified F plasmids into female gametes through sex pili. The conjugation made female gametes return to the state of undifferentiation (called G cells), which meant that they could divide again but were not sexually determined.
After several rounds of cell divisions, F factor and its incompatible plasmid in G cell would seperate. Consequencely, the G cell differentiated into a G+ or G-, which, like zygote, maybe contained genes from both male and female gametes.
Simply, our project included differentiation from mature E. coli to gametes, conjugation between male and female gametes, which resulted in de-differentiation, and sex differentiation through plasmid incompatiblity.
Five systems in our design
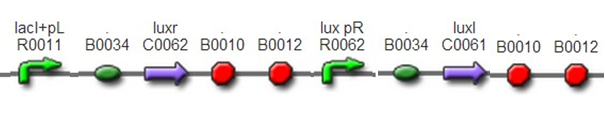
Quorum Sensing systemFunctionalityThis part was designed to sense the density of E. coli and to start gametogenesis at an appropriate time. So , it could imitate the growth and maturity of the animal.
DesignThis part was constructed by assembling standard biobricks: BBa_F2622, BBa_C0261, and terminator BBa_B0015. In this part, luxr was controlled by inducible promoter R0010. IPTG was added to the culture of G+/G- so that this part could work. And IPTG could be washed away to stop quorum sensing before mixing male and female gametes(quorum sensing would disturb conjugation between gametes).
Test
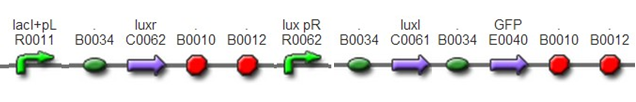
To test this system, we also insert a GFP after LuxI to detect the strength of luxpR and to report the time when cell density reaches the threshold.This was achieved by assembling BBa_F2622, BBa_C0261, and BBa_I13504.
AchievementsWe have constructed above two devices, however, sequence results suggested the whole devices except the first biobrick BBa_F2622 we used was right. Maybe some unknown problems came up when we transformed BBa_F2622 from the distribution kit.
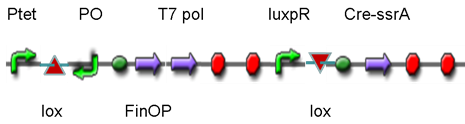
Controlled DNA Invertase device insertted in genomeFunctionalityThis part was a crutial sysytem for our project since it was designed to control the differentiation from mature E.coli to gametes. This device used cre and flpe invertases, and could “invert only once at one time”. Moreover, it is reversible and could invert again after conjugation. Therefore, this part is very suitable for the control of differention and de-differentiation in our project.
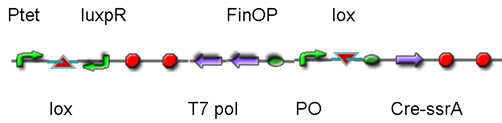
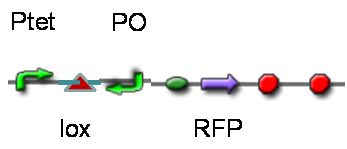
DesignThis part was derived from DNA Invertase Cascade (DIC) Counter (reference:[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2690711/ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2690711/]), and have been redesigned to be used in our system. In this part, there were a reverse Lox site and a reverse PO promoter between Ptet and the RBS, and a forward Lox site after luxpR. PO promoter was from phage (see following) and FinOP is a conjugation inhibition system consisting of an antisense RNA (finP) and a small protein (finO) (reference: https://2009.igem.org/Team:UNICAMP-Brazil).
LuxpR was the quorum sensing promoter, once the cell density of G+/G- reached the threshold, the luxpR promoter would express cre invertase (with an LAA-ssrA degradation tag). Cre invertase would recognize two oppositely-oriented Lox sites, and invert the DNA sequence between them. Due to the inverted orientation of the recombination gene with respect to luxpR promoter, further expression of the recombinase protein ceases and DNA orientation is fixed. (PS: PO promoter didn’t express in G+/G- cells) Inverted DNA sequence: The invertion stopped the expression of FinOP, so that the inhibition of conjugation was eliminated and male E.coli could construct sex pili, which meant that it differentiated into gametes.
Construct : In the part,reverse Lox site and reverse PO promoter were constructed by PCR using PO promoter BBa_I746361 as template. The biobrick suffix was added onto the 5’ end of the upstream primer, Xbal site and reverse Lox site were added onto the 5’ end of the downstream primer. Therefore, the PCR product was “reverse Lox site + reverse PO promoter”. Then, we assembled Ptet with “reverse Lox site + reverse PO promoter” The LAA-ssrA degradation tag of cre-ssrA was chosed among the four ssrA tags with different degeradation rate. The LAA was the fast one. (reference: [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106306/]) To construct RBS-cre-ssrA, BBa_I718008 was used as a template and the LAA tag was added onto the downstream primer.
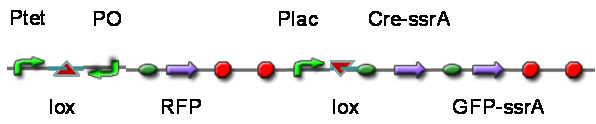
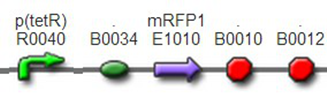
TestTo test the function of above controlled DNA Invertase device, we also intended to constructed the following construct: RFP reporter was put between the two oppositely-oriented promoters, and GFP-ssrA was put after cre-ssrA, Plac was used to replace luxpR. Therefore, we can simply construct this device, and test its function without the need for other systems. We could use IPTG to induce the Plac, and detect the fluorescence of GFP and RFP to test whether the “Controlled DNA Invertase device” works. AchievementsWe have successfully construct Ptet-reverse Lox-reverse PO-RFP([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087016 BBa_K1087016]), and sequence results were good. What’s more, RFP were normally expressed, obsevred and measured. See results: We also build a model about the function of Controlled DNA Invertase device,see model:
Real-time fluorescence reporterFunctionalityReal-time reporters was used to report the changes in E.coli immediately and in time. In our project, GFP and RFP were expressed in G+ and G- respectively. When G+ differentiated into P+ because of the DNA invertion in “Controlled DNA Invertase device”, we need real-time reporters to tell us it’s time to move to the next step.
Construct processSince mature GFP is very stable, we need a GFP with ssrA degradation tag. (reference: [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106306/ http://www.ncbi.nlm.nih.gov/pmc/articles/PMC106306/]). We constructed the real-time GFP reporter [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087013 BBa_K1087013] by assembly four biobricks from distribution kits: BBa_I712074, B0034, BBa_E0044,and teminator BBa_B0015.
We also constructed some other reporters to be used: T7 promoter + constitutive promoter + mRFP1, BBa_K1087020 In this part, the reporter mRFP1 was controlled by both the strong T7 promoter and one weak constitutive promoter BBa_J23117. Thus, it was strongly expressed when cotransfected with T7 polymerase while less expressed alone.
T7 promoter-B0034-amilCP-teminator, BBa_K1087003 AmilCP is a very strong reporter, even can be seen by naked eyes (Reference BBa_K592025, https://2012.igem.org/Team:Groningen/Construct)
Three regulators and its responding promotersFunctionalityIn our project, we need three regulators and their responding promoters to control the expression of target genes. (1)The first regulator was T7 polymerase and T7 promoter.T7 polymerase was put in the “Controlled DNA Invertase device”. In G+ cells, T7 polymerase was in the right orientation and expressed by Ptet. Threefore, Pt7 could express real-time reporters. When T7 polymerase sequence was inverted in P+ cells, further expression of real-time would cease.
(2)The second regulator was riboregulator.(Reference: https://2006.igem.org/wiki/index.php/Berkeley2006-RiboregulatorsMain) Functionality: In our design, BBa_K145215 (“TetR promoter + J23066 RiboKey”) was put in G- on the plasmid incompatible with F factor.
The Ribolock “TetR promoter + lock3d” was constructed and put in G+ on the F plasmid.
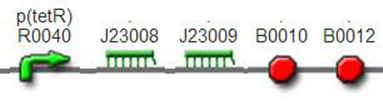
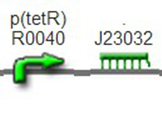
We improved an existing biobrick part BBa_I714070([pTet][Lock3]). We could not transforme this part from the distribution kit, and the QC results for this part in the registry were bad, too. Bad sequence, inconsistent resistence, and bad gel results. Therefore, we constructed this part by ourselves. We assembled two basic parts Ptet BBa_R0040 and [lock3d] BBa_J23032 to create a new part: BBa_K1087004 Sequence results were confired the successful construclion, and we did lots of tests to prove its function.
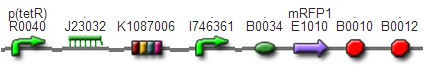
We assembled BBa_K1087004 with mRFP to create [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K1087015], and we ligate [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K1087015] with RiboKey BBa_K145215 to create the new [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087021 BBa_K1087021]. [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K1087015] [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087021 BBa_K1087021]
Negetive control: BBa_J61046 We transformed [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K1087015], [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087021 BBa_K1087021] into E.coli DH5α, respectively. And we used Ptet + mRFP1 (BBa_I13521) as positive control, irrelevant BBa_J61046 as negative control. The transformed cells were cultured separately in the same condition and measured the intensity of fluorescence at the same time. See results.
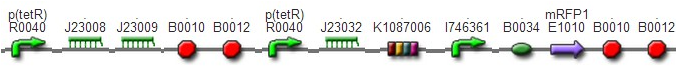
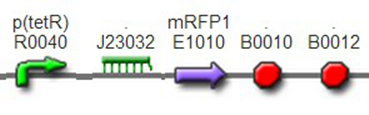
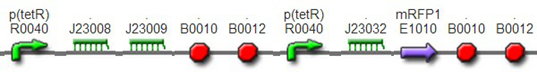
(3)The third regulator was delta activator from phiR73 phage BBa_I746352The third regulator was delta activator from phiR73 phage BBa_I746352, which can induce strongly PO promoter BBa_I746361 from P2 phage. (Reference: https://2009.igem.org/Team:Cambridge/Project/Amplification/Characterisation). Test We also built some bigger constructs to test the function of Riboregulator and PhiR73 activator . We ligated PO promoter with mRFP1 to create [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K1087018] so that we can test the basal level of PO promoter. We also ligated “TetR promoter + lock3d” with phiR73 activator to create [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087007 BBa_K1087007]. Then, we assembled [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087007 BBa_K1087007] with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K1087018] to create [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087022 BBa_K1087022].
[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K1087018]
[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087022 BBa_K1087022] [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087022 BBa_K1087023]
F factor and its incompatible plasmidFunctionalityF plasmid was intended to be constructed in G+, and would be transferred to P- when conugation.The RiboKey on F plasmid would be open the Ribolock on the plasmid in P-.Then ,the phiR73 activator would be expressed and induce PO promoter on the “Controlled DNA Invertase device”. Finally,the DNA sequence between Lox sites would be inverted to the original state. Cells returned to the state of un-differentiation (called G cells).
DesignWe intended to modify F palsmid using RED sysytem (reference:[http://www.ncbi.nlm.nih.gov/pubmed/10829079]).
And we intended to make use of the stardard biobrick plasmids (reference:[1]) that were incompatible with F Factor.
ResultResult 1 Test the function of Riboregulator
We assembled BBa_K1087004 with mRFP to create [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K1087015], band we assembled [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K1087015] with RiboKey BBa_K145215 to create [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087021 BBa_K1087021]. [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K1087015] [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087021 BBa_K1087021]
Negative control: BBa_J61046
We transformed [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K1087015] and [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087021 BBa_K1087021] into E. coli DH5, respectively. And we used Ptet+mRFP1 (BBa_I13521) as positive control, irrelevant BBa_J61046 with no fluorescence gene as negative control.
All of the constructs were expressed in high copy plasmid PSB1X3. The transformed cells were cultured in 37℃, 200rpm.
We used fluorescence microscope to observe the fluorescence at 16h, and we also used Thermo Varioskan Flash to quantitatively measure the fluorescence intensity at 16h, 18h, and 19h, respectively. 150ul culture fluid was added into each well of the 96-well black plate to measure the fluorescence intensity, and OD600 were measured using spectrophotometer at the same time one by one to quantify the concentration of bacteria. Each sample had three duplicates when cultured. And the fluorescence value was handled through mathematical method. The fluorescence intensity of above transformants was divided by the OD600 value and minus that of negative control. Data Qualitative representation of Riboregulator 1, color change with time revealed the expression level of corresponding parts.
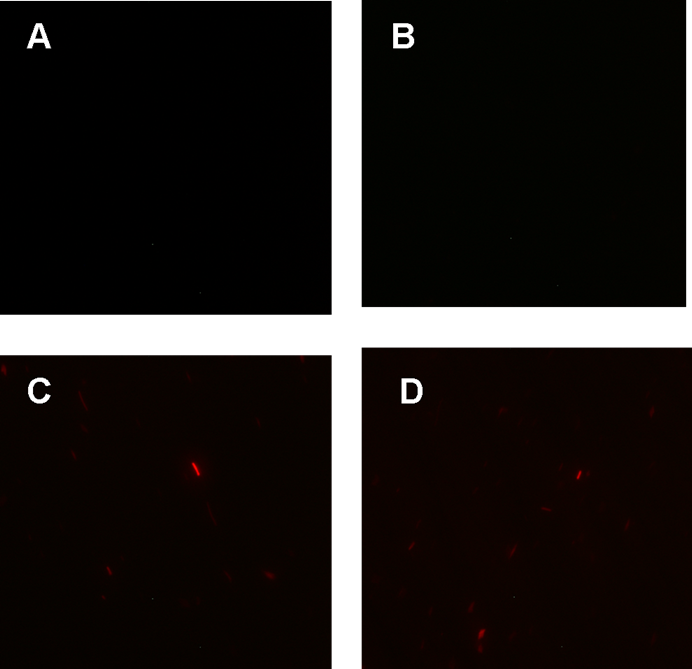
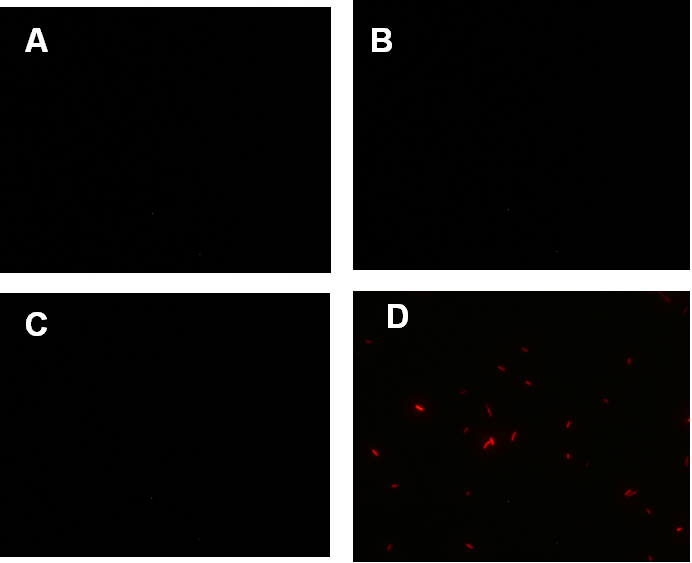
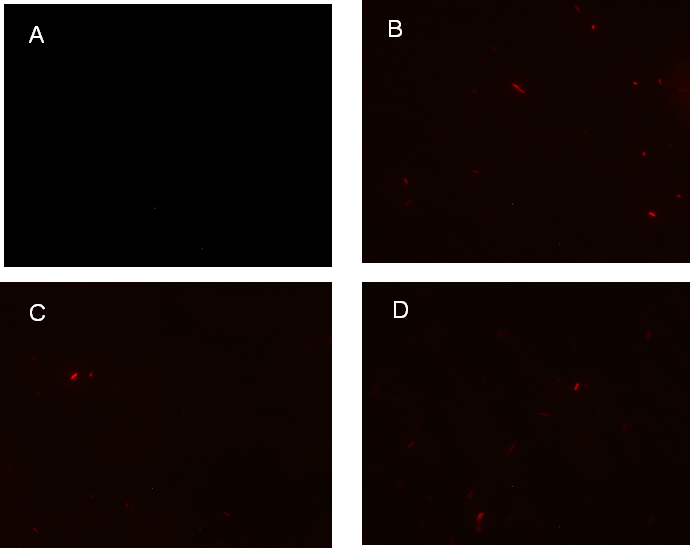
Figure 1. fluorescence observation using fluorescence microscope. A. Negative control: Coletit DH5αtransformed with B Ba_J61046 in PSB1X3. B. Coletit DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K1087015]. C. Coletit DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K10870]21. D. Positive control: Coletit DH5αtransformed with B Ba_I13521.
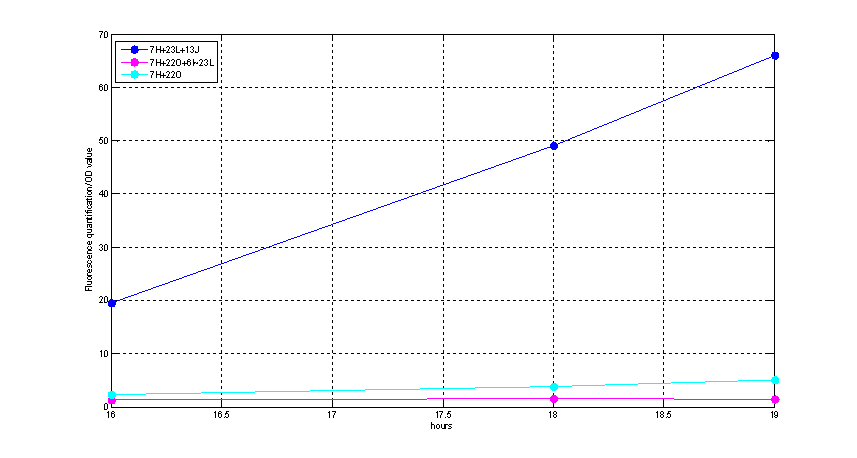
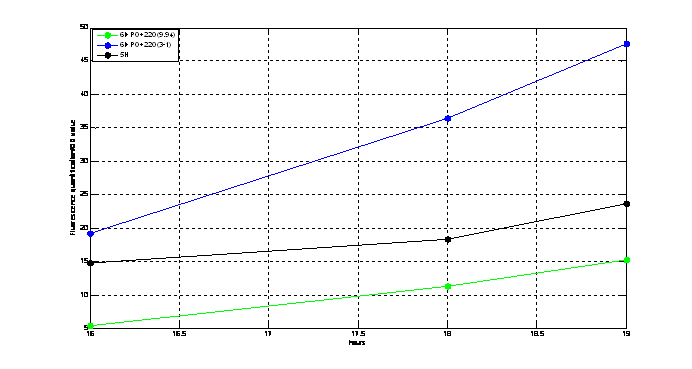
Figure2. quantitatively measurement of the fluorescence intensity. Red: Coletit DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K1087015]. Blue: Coletit DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K10870]21. Black: Positive control: Coletit DH5αtransformed with B Ba_I13521. PS: the fluorescence intensity of above transformants was divided by the OD600 value and minus that of negative control.
According to above figures, it’s obvious that the ribolock BBa_K1087004 worked well, and its fluorescence intensity was only 2.6531% compared to positive control even at 19h, while that of lock & key ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087021 BBa_K1087021]) was 91.7138% at 19h. The results suggested that the key BBa_K145215 could hugely improve the expression level of the lock BBa_K1087004.
Data:
Result 2 Test the function of the Riboregulator and PhiR73 activator together
We assembled PO promoter with mRFP1 to create [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K1087018],so that we can test the basal expression level of PO promoter. We also assembled “TetR promoter + lock3d” with phiR73 activator to create [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087007 BBa_K1087007]. Then, we assembly [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087007 BBa_K1087007] with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K1087018] to create [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087022 BBa_K1087022]. Finally, we assembled RiboKey BBa_K145215 with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087022 BBa_K1087022] to create [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087022 BBa_K108702]3.
[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K1087018]
[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087022 BBa_K1087022]
[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087022 BBa_K108702]3 We transformed [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K1087018], [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087022 BBa_K1087022], and [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087022 BBa_K108702]3 into E. coli DH5α, respectively. And we used irrelevant BBa _J61046 with no fluorescence gene as negative control. The transformed cells were cultured separately in the same condition and measured fluorescence intensity at same time.
The measurement was done mainly the same as result 1. Data Qualitative representation of Riboregulator 2, color change with time revealed the expression level of corresponding parts.
Figure 1. Figure 1. fluorescence observation using fluorescence microscope. A. Negative control: Ecoli DH5αtransformed with BBa_J61046 in PSB1X3. B. E. coli DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K1087018]. C. E. coli DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K10870]22. D. E. coli DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K10870]23.
Figure2. Quantitatively measurement of the fluorescence intensity. Blue: E. coli DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K10870]23. Green: E. coli DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087015 BBa_K10870]18. Pink: E. coli DH5αtransformed with BBa_K108722. PS: the fluorescence intensity of above transformants was divided by the OD600 value and minus that of negative control.
We could see clearly from above figures that the basal expression level of PO promoter was really low, and its strength could be improved about 11 times when induced by PhiR73 controlled by ribokey and lock. Data:
Result 3 Test the function of Part BBa_K1087016 BBa_K1087016Normally, we picked two colonies from the agar plate for [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K108701]6, called A and B. We transformed [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K108701]6A and B into E. coli DH5, respectively. And we use Ptet+mRFP1(BBa_I13521) as positive control, irrelevant BBa_J61046 with no fluorescence gene as negative control. The transformed cells were cultured separately in the same condition and measured fluorescence intensity at same time.
The measurement was done mainly the same as result 1.
Figure1. fluorescence observation using fluorescence microscope. A. Negative control: E. coli DH5αtransformed with BBa_J61046 in PSB1X3. B. E. coli DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K108701]6A. C. E. coli DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K108701]6B. D. Positive control: E. coli DH5αtransformed with BBa_I13521.
Figure2. Quantitatively measurement of the fluorescence intensity. Blue: E. coli DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K108701]6A. Black: Positive control, E. coli DH5αtransformed with BBa_I13521. Green: E. coli DH5αtransformed with [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K108701]6B. PS: the fluorescence intensity of above transformants was divided by the OD600 value and minus that of negative control.
From above figures, we could conclude that the Ptet and mRFP1 in [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K108701]6 worked well. It was comparable to even stronger than the positive control BBa_I13521 in fluorescence intensity. For [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K108701]6A, it expressed about twice fluorescence of BBa_I13521. Results for sequencing suggested that the first half of the Ptet was lost in [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1087018 BBa_K108701]6A, which may account for its increase in strength.
Data:
|
 "
"