Team:Tokyo Tech/Project/M13 supplement
From 2013.igem.org
Tryalnigro (Talk | contribs) |
|||
| (4 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<div id="text-area"><br> | <div id="text-area"><br> | ||
<div class="box" id="title"> | <div class="box" id="title"> | ||
| - | <p style="line-height:0em; text-indent:0em;">Supplement to M13 "shuriken" project</p> | + | <p style="line-height:0em; text-indent:0em;" name="top">Supplement to M13 "shuriken" project</p> |
</div> | </div> | ||
<div class="box"> | <div class="box"> | ||
| - | <h1>M13 plasmid construction</h1> | + | <h1>1. M13 plasmid construction</h1> |
<h2> | <h2> | ||
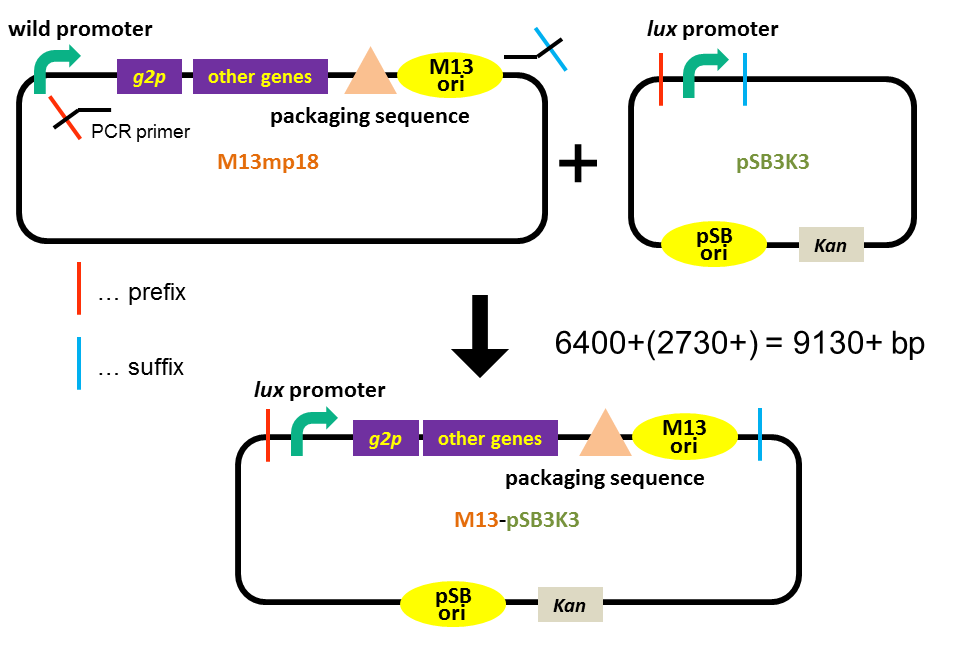
<p>In order to construct a plasmid that enables inducible release of M13 phage and can be transported by a phage particle, we combined M13 genome lacking <i>g2p</i> promoter (derived from M13mp18 phage vector), pSB3K3 backbone and <i>lux</i> promoter. The plasmid has two different types of replication origins: M13 origin and pSB origin (p15A). The size of the plasmid is 9130 bp (Fig. 3-8-1). | <p>In order to construct a plasmid that enables inducible release of M13 phage and can be transported by a phage particle, we combined M13 genome lacking <i>g2p</i> promoter (derived from M13mp18 phage vector), pSB3K3 backbone and <i>lux</i> promoter. The plasmid has two different types of replication origins: M13 origin and pSB origin (p15A). The size of the plasmid is 9130 bp (Fig. 3-8-1). | ||
| Line 12: | Line 12: | ||
</h2> | </h2> | ||
| - | <h1>Replication machinery of M13 origin</h1> | + | <h1>2. Replication machinery of M13 origin</h1> |
<h2> | <h2> | ||
<p>M13 origin is composed of four hairpin loops of 177 nucleotides of a single stranded DNA. The hairpin is divided to two areas of different functions: - strand origin and + strand origin (Fig. 3-8-2). - strand origin is needed for a + strand DNA to be a double stranded DNA. Besides, + strand origin is needed for production of a + strand DNA from a double stranded DNA. - strand DNA is synthesized by host’s RNA polymerase and DNA polymerase. + strand synthesis is triggered by G2p nickase, M13 plasmid-coded protein. The new synthesized + strand DNA can be double stranded DNA again. | <p>M13 origin is composed of four hairpin loops of 177 nucleotides of a single stranded DNA. The hairpin is divided to two areas of different functions: - strand origin and + strand origin (Fig. 3-8-2). - strand origin is needed for a + strand DNA to be a double stranded DNA. Besides, + strand origin is needed for production of a + strand DNA from a double stranded DNA. - strand DNA is synthesized by host’s RNA polymerase and DNA polymerase. + strand synthesis is triggered by G2p nickase, M13 plasmid-coded protein. The new synthesized + strand DNA can be double stranded DNA again. | ||
| Line 18: | Line 18: | ||
[[Image:PSB-M13_Phage_replication_Fig.2.PNG|600px|thumb|center|Fig. 3-8-2. Functioin of M13 origin]] | [[Image:PSB-M13_Phage_replication_Fig.2.PNG|600px|thumb|center|Fig. 3-8-2. Functioin of M13 origin]] | ||
</h2> | </h2> | ||
| - | <h1>Phage release regulation</h1> | + | <h1>3. Phage release regulation</h1> |
<h2> | <h2> | ||
| - | <p>+ strand DNA has another function; it can assembly a phage particle. To regulate release of a phage particle, we altered the promoter upstream of <i>g2p</i> to <i>lux</i> promoter, which is an AHL-inducible promoter. | + | <p>+ strand DNA has another function; it can assembly a phage particle. To regulate release of a phage particle, we altered the promoter upstream of <i>g2p</i> to <i>lux</i> promoter, which is an AHL-inducible promoter. With M13 origin only, when the promoter is repressed, not only phage release but also DNA replication are inhibited (Fig. 3-8-3A). To solve this problem, we added pSB origin (Fig. 3-8-3B). As a result, M13 origin and <i>lux</i> promoter can be used as a trigger of phage release because pSB origin amplifies DNA. |
</p></h2> | </p></h2> | ||
<gallery widths="400px" heights="300px" style="margin-left: auto; margin-right: auto;"> | <gallery widths="400px" heights="300px" style="margin-left: auto; margin-right: auto;"> | ||
| Line 27: | Line 27: | ||
</gallery> | </gallery> | ||
| - | <h1>Phage assembly</h1> | + | <h1>4. Phage assembly</h1> |
<h2> | <h2> | ||
| - | <p>A hairpin loop of a single stranded DNA (+ strand) is necessary for M13 phage assembly. The sequence that forms hairpin loop is called “packaging sequence”. G5p, a single-strand binding protein, protects the single stranded DNA (Fig. 3-8-4A). Coat proteins and pore proteins are all embedded in cell membranes before assembly. The phage particle is primarily assembled from G7p and G9p, two types of coat proteins, which act on the hairpin loop. In the course of the assembly, G5p proteins are replaced with G8p proteins, which form the major part of a phage particle (Fig. 3-8-4B). | + | <p>A hairpin loop of a single stranded DNA (+ strand) is necessary for M13 phage assembly. The sequence that forms hairpin loop is called “packaging sequence”. G5p, a single-strand binding protein, protects the single stranded DNA (Fig. 3-8-4A) from degradation or being double stranded. Coat proteins and pore proteins are all embedded in cell membranes before assembly. The phage particle is primarily assembled from G7p and G9p, two types of coat proteins, which act on the hairpin loop. In the course of the assembly, G5p proteins are replaced with G8p proteins, which form the major part of a phage particle (Fig. 3-8-4B). |
</p> | </p> | ||
[[Image:Titech2013_M13_phage_assembly_and_infection_Fig.1.PNG|600px|thumb|center|Fig. 3-8-4A. The start of phage assembly]] | [[Image:Titech2013_M13_phage_assembly_and_infection_Fig.1.PNG|600px|thumb|center|Fig. 3-8-4A. The start of phage assembly]] | ||
| Line 35: | Line 35: | ||
</h2> | </h2> | ||
| - | <h1>Phage infection</h1> | + | <h1>5. Phage infection</h1> |
<h2> | <h2> | ||
<p>M13 phage infects only F+ strains, which possess F pili. First, G3p, one of the coat proteins, binds to an F pilus (Fig. 3-8-5). Second, the F pilus contracts; the phage approaches the host’s membrane. Finally, one of the domains of G3p binds to TolA, one of the host’s membrane proteins. | <p>M13 phage infects only F+ strains, which possess F pili. First, G3p, one of the coat proteins, binds to an F pilus (Fig. 3-8-5). Second, the F pilus contracts; the phage approaches the host’s membrane. Finally, one of the domains of G3p binds to TolA, one of the host’s membrane proteins. | ||
</p> | </p> | ||
| - | [[Image:Titech2013_M13_phage_assembly_and_infection_Fig.3.PNG|600px|thumb|center|Fig. 3-8-5. | + | [[Image:Titech2013_M13_phage_assembly_and_infection_Fig.3.PNG|600px|thumb|center|Fig. 3-8-5. Function of M13 origin]] |
</h2> | </h2> | ||
| - | <h1>References</h1> | + | <h1>6. References</h1> |
<h2> | <h2> | ||
| - | < | + | <ol> |
| - | < | + | <li>Model, P. & Russel, M. (1988) <i>in</i> The Bacteriophages (ed. Calendr, R.), 2, pp.375-456, Plenum. |
| - | < | + | <li>Horiuchi, K. (1990) <i>Jpn. J. Genet.</i>, <b>65</b>, 225-241. |
| - | < | + | <li>Wickner, W. & Brutlag, D. & Scheckman, R. & Kornberg, A. (1972) <i>Proc. Natl. Acad. Sci. USA</i>, <b>69</b>, 965-969. |
| - | < | + | <li>Geider, K. & Kornberg, A. (1974) <i>J. BIol. Chem.</i>, <b>249</b>, 3999-4005. |
| - | < | + | <li>Horiuchi, K. & Zinder, N.D. (1976) <i>Proc. Natl. Acad. Sci. USA</i>, <b>73</b>, 2341-234. |
| - | < | + | <li>Gray, C.P. & Sommer, R. & Polke, C. & Beck, E & Schaller, H. (1978) <i>Proc. Natl. Acad. Sci. USA</i>, <b>75</b>, 50-53. |
| - | < | + | <li>Meyer, T.F. & Geider, K. & Kruz, C. & Schaller, H. :<i>Nature</i>. <b>278</b>, 365-367 (1979) |
| - | < | + | <li>Marvin, D.A. & Hohn, B. (1969) <i>Bacteriol. Rev.</i>, <b>33</b>, 172-209. |
| - | < | + | <li>Rasched, I. & Oberer, E. (1986) <i>Microbiol. Rev.</i>, <b>50</b>, 401-427. |
| - | < | + | <li>Eckert, B. & Schmid, F.X. (2007) <i>J. Mol. Biol.</i>, <b>373</b>, 452-461. |
| + | <li>Houbiers, M.C. & Hemminga, M.A. (2004) <i>Mol. Membr. Biol.</i>, <b>21</b>, 351-359. | ||
| + | </ol> | ||
</h2> | </h2> | ||
</div><br> | </div><br> | ||
| + | <html><div align="center"><a href="https://2013.igem.org/Team:Tokyo_Tech/Project/M13_supplement#top"><img src="https://static.igem.org/mediawiki/2013/f/f0/Titeh2013_backtotop.png" width="200px"></a></div></html> | ||
</div> | </div> | ||
Latest revision as of 04:18, 27 October 2013
Supplement to M13 "shuriken" project
Contents |
1. M13 plasmid construction
In order to construct a plasmid that enables inducible release of M13 phage and can be transported by a phage particle, we combined M13 genome lacking g2p promoter (derived from M13mp18 phage vector), pSB3K3 backbone and lux promoter. The plasmid has two different types of replication origins: M13 origin and pSB origin (p15A). The size of the plasmid is 9130 bp (Fig. 3-8-1).
2. Replication machinery of M13 origin
M13 origin is composed of four hairpin loops of 177 nucleotides of a single stranded DNA. The hairpin is divided to two areas of different functions: - strand origin and + strand origin (Fig. 3-8-2). - strand origin is needed for a + strand DNA to be a double stranded DNA. Besides, + strand origin is needed for production of a + strand DNA from a double stranded DNA. - strand DNA is synthesized by host’s RNA polymerase and DNA polymerase. + strand synthesis is triggered by G2p nickase, M13 plasmid-coded protein. The new synthesized + strand DNA can be double stranded DNA again.
3. Phage release regulation
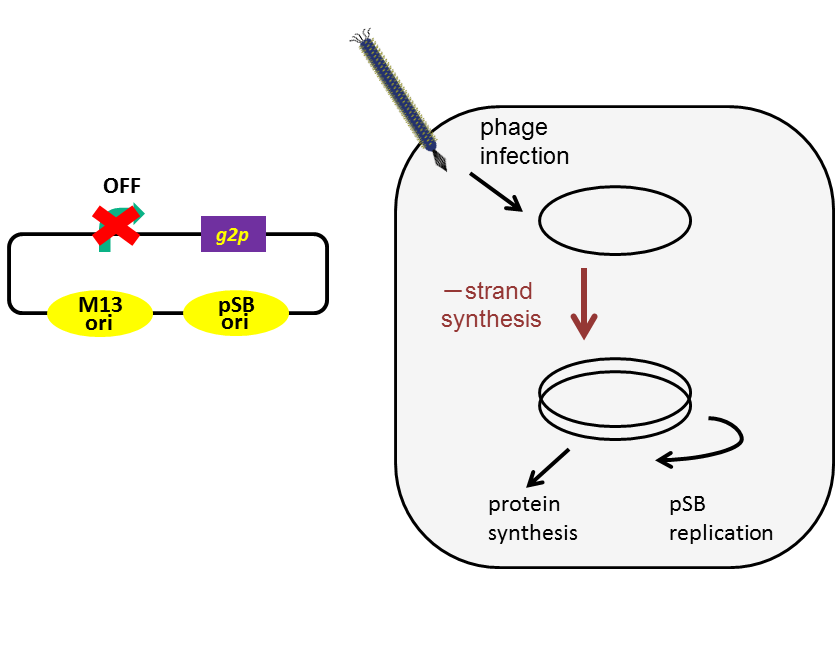
+ strand DNA has another function; it can assembly a phage particle. To regulate release of a phage particle, we altered the promoter upstream of g2p to lux promoter, which is an AHL-inducible promoter. With M13 origin only, when the promoter is repressed, not only phage release but also DNA replication are inhibited (Fig. 3-8-3A). To solve this problem, we added pSB origin (Fig. 3-8-3B). As a result, M13 origin and lux promoter can be used as a trigger of phage release because pSB origin amplifies DNA.
4. Phage assembly
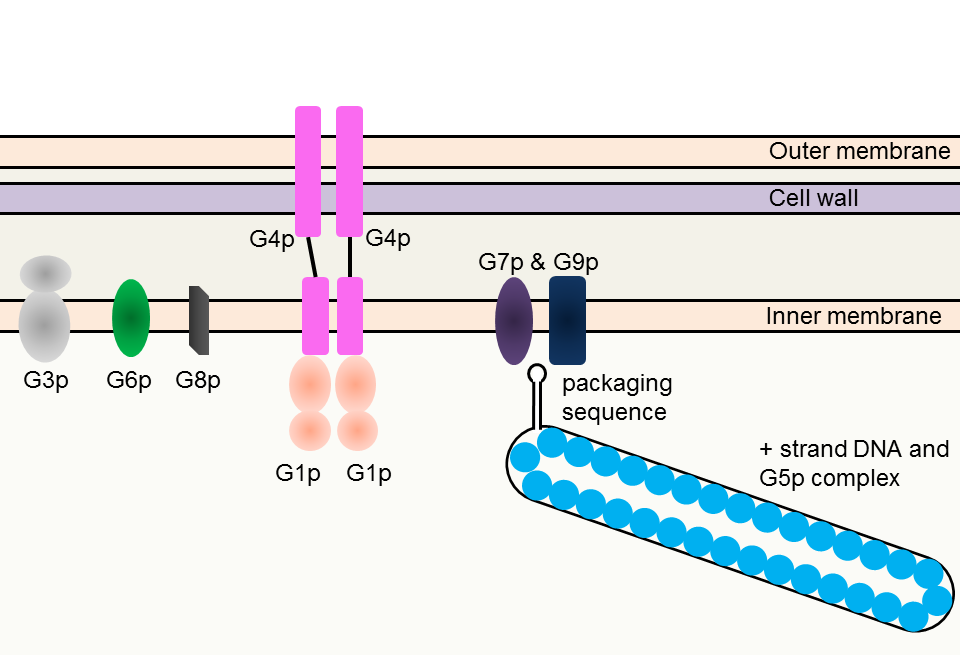
A hairpin loop of a single stranded DNA (+ strand) is necessary for M13 phage assembly. The sequence that forms hairpin loop is called “packaging sequence”. G5p, a single-strand binding protein, protects the single stranded DNA (Fig. 3-8-4A) from degradation or being double stranded. Coat proteins and pore proteins are all embedded in cell membranes before assembly. The phage particle is primarily assembled from G7p and G9p, two types of coat proteins, which act on the hairpin loop. In the course of the assembly, G5p proteins are replaced with G8p proteins, which form the major part of a phage particle (Fig. 3-8-4B).
5. Phage infection
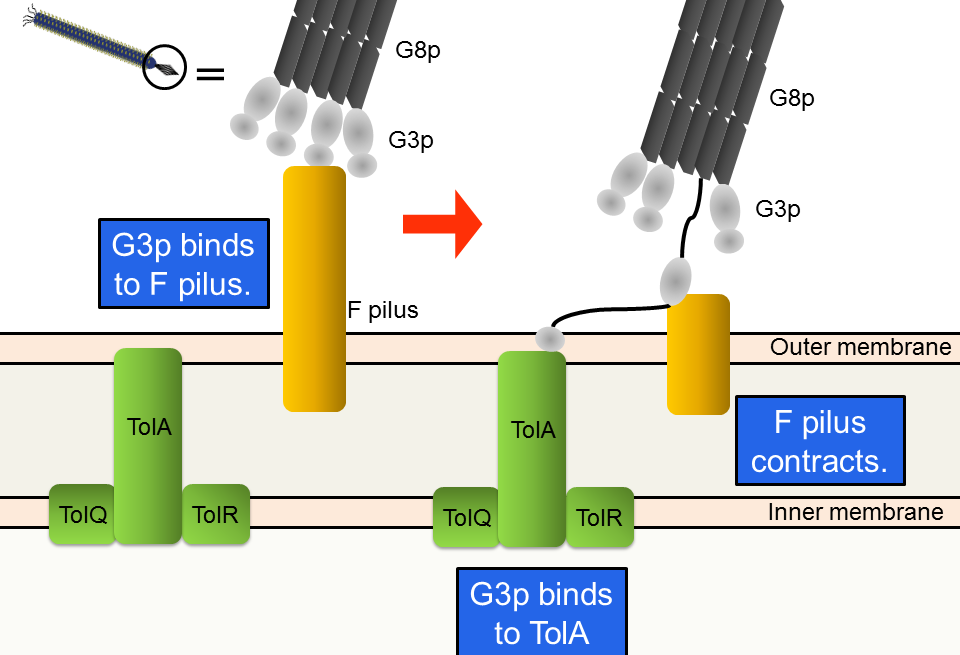
M13 phage infects only F+ strains, which possess F pili. First, G3p, one of the coat proteins, binds to an F pilus (Fig. 3-8-5). Second, the F pilus contracts; the phage approaches the host’s membrane. Finally, one of the domains of G3p binds to TolA, one of the host’s membrane proteins.
 "
"