From 2013.igem.org
Ran the gel from the 26/07/2013 again for 45min

Digestion of plasmid backbone (pSB1C3)
- Three different reactions
- Master Mix (changes were made from the protocol)
- 5ul NEB Buffer 3.1
- 0.5ul of EcoRI
- 0.5ul of PstI
- 19ul of dH2O
- For each reaction add:
- 4ul of linearized backbone
- 4ul of enzyme master mix
- digest at 37C for 30min
- heat kill at 80C for 20min
Digestion of the limonene biobrick plasmid backbone (BBa_K118025)
- Using clone 1.1 purified plasmid DNA
- For the reaction adding:
- 5ul NEB Buffer 3.1
- 2ul of EcoRI
- 2ul of PstI
- 2.5ul of dH2O
- 38.5ul of DNA
- digest at 37C for 30min
- heat kill at 80C for 20min
Ligation of insert to vector
- For this experiment, two separate samples will be run; a control and the actual ligation.
- Calculate appropriate vector:insert ratio and convert to a 1:1 ratio
- Add the following substances
- 11.1ul of dH2O
- 1ul of QS Ligase *
- 5ul of 4xQS Buffer (vortex before use)
- Mix thoroughly by pipetting
- Incubate at room temperature for 5 min to create cohesive ends
- Run 2.5-5ul of the ligation mixture onto an agarose gel to check ligation efficiency against a known marker. Add 1ul of dye to aid the visualization of results.
- Transform it into competent cells (E.coli)
*For the control, no QS Ligase is added but its equivalent volume (1ul) is made up by dH2O.
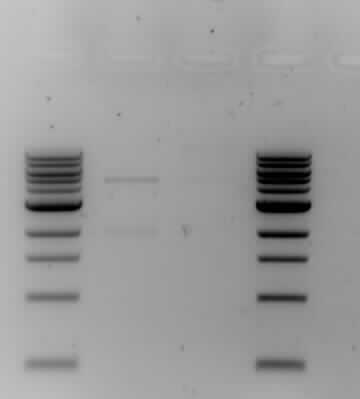
- The results from the gel electrophoresis show four lanes; two 1kb markers on either end (the first and forth lane), one control (the second lane) and the actual ligation results (the third lane). The control has two bands which shows that it has not been ligated, one band is around the 2kb area which is lightly-staining and one band is heavy-staining around the 5kb area. The actual ligation shows a lightly-staining smear, conclusive with the formation of a plasmid. The results are sufficiently good to carry on with the next phase of the experiment - the transformation.
 "
"

