|
7/5/13
Bryan did extra research into the best protocol to get the results that we are hoping for. He found a paper entitled, "Genetic Control of Capsid Length in Bacteriophage T4" By:A. H. DOERMANN, F. A. EISERLING, AND LINDE BOEHNER
7/8/13
Today we need to address some problems we ran into and make some plans for the future.
1. Plaque sizes aren't consistent, even though we are using the same agarose each time.
2. We haven't been able to observe plaque phenotypes staying the same through multiple generations
3. We have tried multiple liquid cultures and rounds of mutagenesis but we haven't been able to observe clearage in a flask yet.
Good things to model
- Change in plaque size vs. agarose concentration
- Mutant vs wild type phage under UV - graph survival rates
7/10/13
Today we are trying out a different but similar protocol. We are hoping to see clearage in our overnight cultures which we haven't seen so far. This new protocol describes that the phage will super infect the bacteria this would explain why our phage wasn't clearing the flask. The paper states,
- "After one round of infection and lysis,all remaining cells become infected. Some of these
- lyse on schedule, liberating phage which superinfect the majority of cells which are still unlysed. This
- superinfection causes inhibition of lysis (9). Starting at 150 min, samples (5 ml) are removed periodically
- and shaken with chloroform to test for the degree of lysis which can be induced. When that test shows that
- most of the cells can be induced to lyse so that clearing is nearly complete (usually at about 180 min
- after initial infection),the culture is chilled in an ice bath and the pregnant bacteria are pelleted by
- centrifugation at about 5,000 rpm for 10 min in a cold centrifuge."
We inoculated a 250 uL flask with 25 mL of LB broth, 100 uL (20 ug/mL) of adenine, and a loop of E. coli. We inoculated 25 mL of M9 with the same ingredients. We let these flasks incubate for ~2 hours until moderately turbid.
After the bacteria is moderately turbid, we added another 100 uL of adenine and 200ul (20 ug/mL) uracil to the 25ml flasks of E. coli B in LB and in M9 along with 500 ul (200 ug/mL) mutagen 5-bromo 2-deoxyuridine and 500 ul T4 phage (1x10^9 pfu/mL) and put that in the shaker to determine if the flask will clear in about three hours.
After 150 minutes, we removed 5 mL of bacterial culture and placed it in a 15 mL tube with 1 mL of chloroform. The LB culture cleared very quickly, while the M9 culture did not clear very well. At 180 minutes, the LB cleared even better and the M9 cleared slightly better. Due to time constraints, we decided to harvest the pregnant bacteria at 180 minutes. We pelleted the bacteria in a centrifuge at 3500 rpm for 10 minutes and resuspended each pellet in 2 mL of broth. We transferred it to a 15 mL tube and added 10 uL of nuclease mix, 5 uL of 1 M MgCl2, and 2 mL of chloroform. We mixed it gently and let it incubate for 30 minutes. Then, we centrifuged it at 2700 rpm for 10 minutes. The supernatant was removed and transferred to a new 15 mL tube.

7/12/13
Today we did a titer on:
LB and M9+ Supernatant
LB and M9+ Lysis inhibited pellet
Re-titer of Mutagenesis 3 and 4
7/15/13

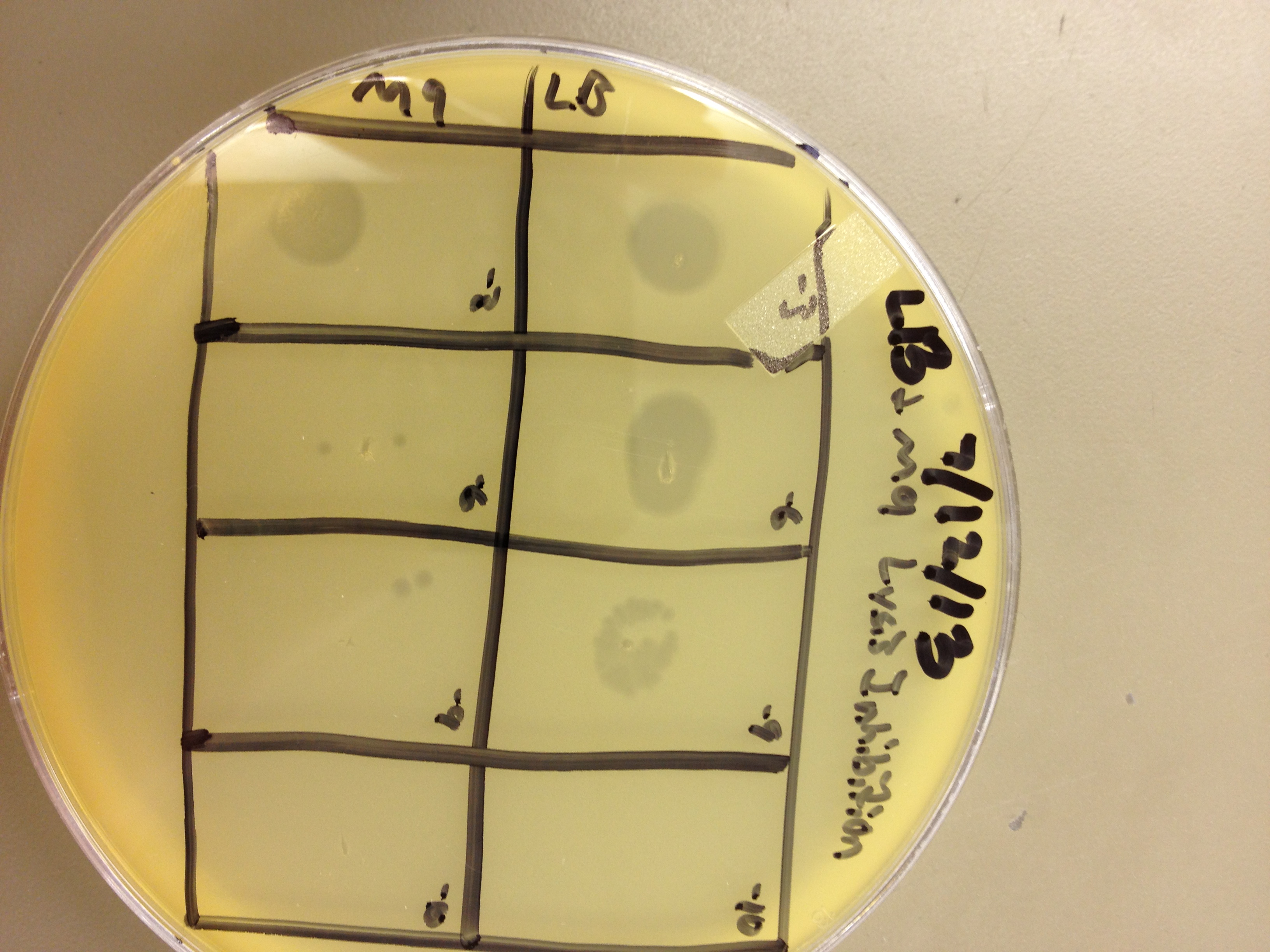
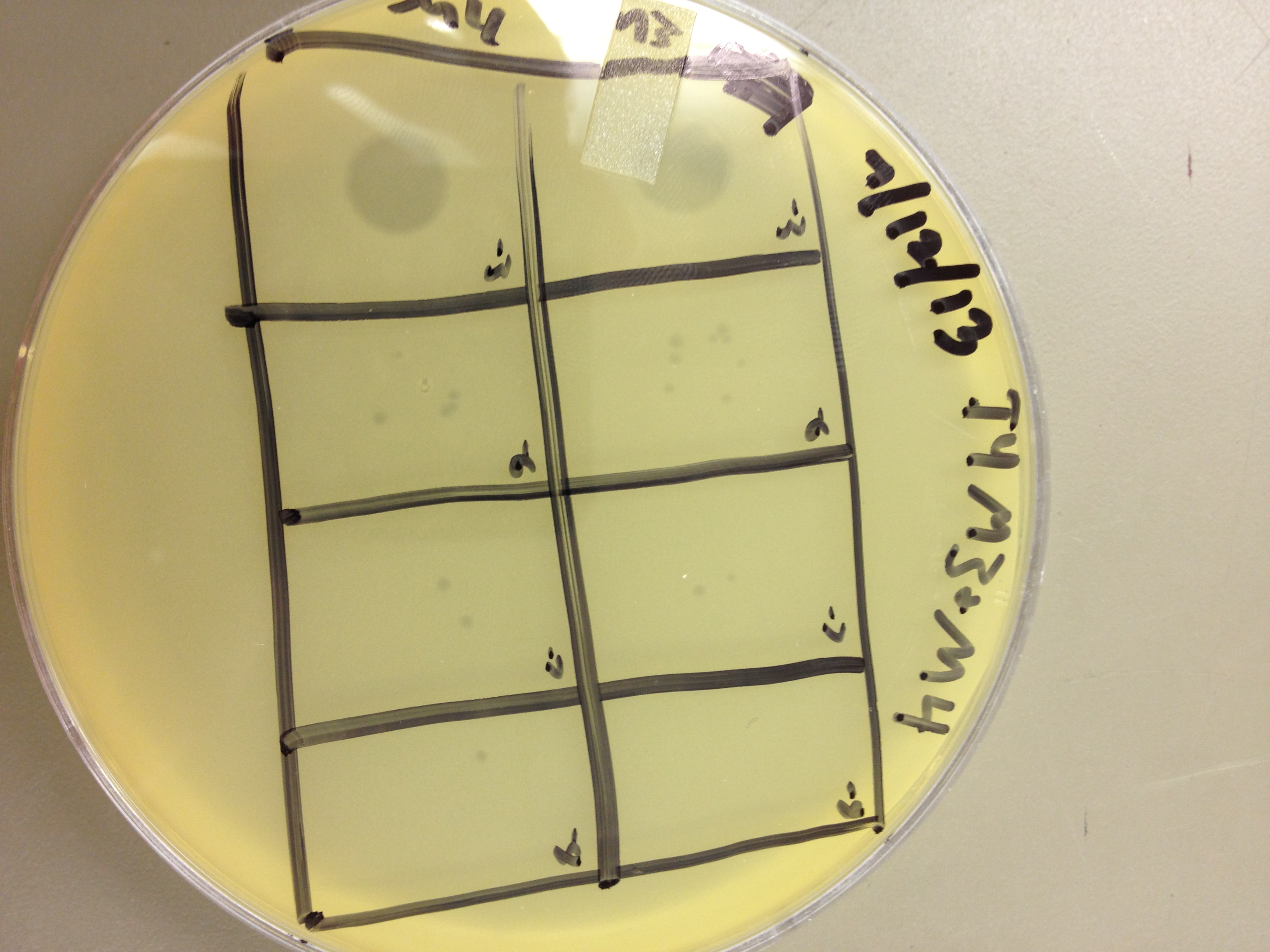
These results gave us some very useful data. We saw that LB broth worked better for growing phage than M9 in this experiment. By comparing the supernatants of the liquid cultures, LB showed one plaque at 10^-8, and M9 showed zero plaques below 10^-6. However, the lysed bacterial pellet for LB had many plaques at 10^-9, while the lysed M9 pellet had 2 plaques at 10^-9. This shows that there are more than 10 times the amount of phage inside the bacteria than outside. This is good, because any phage inside the bacteria are progeny phage which must have been exposed to the mutagen as they were assembling.
Next, we will run the phage through a sucrose or CsCl to try to isolate fractions of large phage and image them.
We also need to refine our EM prep technique.
|