Team:DTU-Denmark/Notebook/8 August 2013
From 2013.igem.org
8 August 2013
Contents |
Lab 208
Main purpose
- PCR for AMO with USER endings.
- Extraction PCR of Nir1
Who was in the lab
Kristian, Julia, Henrike
Procedure
PCR for AMO with USER endings
primers: 17a, 17b
template: gel purified AMO extraction fragment
Program: Standard with 54C annealing temperature and 3:00 extension time
Reaction Mix
| component(per reaction) | without additives | using 5% DMSO | using 1M Betaine |
|---|---|---|---|
| dNTPs | 1uL | 1uL | 1uL |
| HF buffer | 10uL | 10uL | 10uL |
| X7 polymerase | 0.5uL | 0.5uL | 0.5uL |
| MilliQ water | 31.5uL | 29uL | 21.5uL |
| template | 1uL | 1uL | 1uL |
| FW primer | 3uL | 3uL | 3uL |
| RV primer | 3uL | 3uL | 3uL |
| DMSO | - | 2.5uL | - |
| Betaine | - | - | 10uL |
PCR for extraction of Nir1
primers: 41a, 41b
template: colony from plate. One colony was dissolved in 100 uL of MilliQ and 1 uL of this solution was taken as template
program: Touchdown PCR analog to program used on 01-08-2013 to amplify Nir.
Reaction Mix
| component(per reaction) | without additives | using 5% DMSO | using 1M Betaine |
|---|---|---|---|
| dNTPs | 1uL | 1uL | 1uL |
| HF buffer | 10uL | 10uL | 10uL |
| X7 polymerase | 0.5uL | 0.5uL | 0.5uL |
| MilliQ water | 31.5uL | 29uL | 21.5uL |
| template | 1uL | 1uL | 1uL |
| FW primer | 3uL | 3uL | 3uL |
| RV primer | 3uL | 3uL | 3uL |
| DMSO | - | 2.5uL | - |
| Betaine | - | - | 10uL |
Comment
Both PCRs mentioned above were successful when DMSO was added.
Results
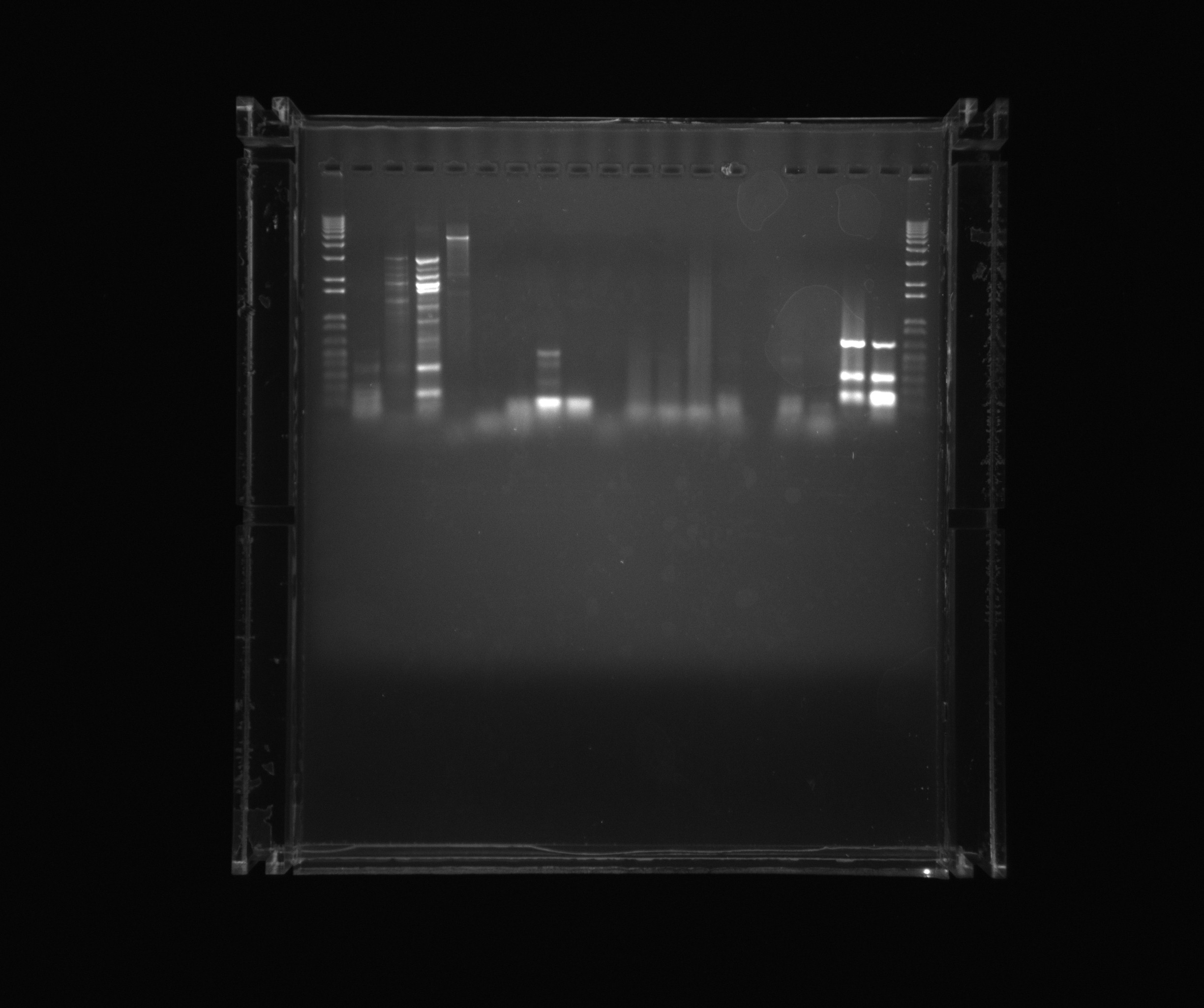
Gel on yesterdays PCR
- 1kb ladder
- Yesterdays sample 1
- Yesterdays sample 2
- Yesterdays sample 3
- Yesterdays sample 4
- Neg.
- Neg.
- Nir2
- cycAX Histag
- 1kb ladder
Gel on ON PCR samples
- 1kb ladder
- Nir1
- Nir1 5%DMSO
- Nir1 1M Betaine
- Nir1 3M Betaine
- Nir2 3M Betaine U
- Nir2 1M Betaine U
- Nir2 5%DMSO U
- Nir2 U
- Neg (Nir)
- Ref 0%DMSO
- Ref 2%DMSO
- Ref 5%DMSO
- AMO U
- AMO 2%DMSO U
- Neg (AMO)
- NirG 5%DMSO
- NirG
- 1kb ladder
Gel on todays PCR samples
- 1kb ladder
- Nir Q5 poly
- Nir 2%DMSO Q5 poly
- Nir 3M Betaine Q5 poly
- Nir 5%DMSO Q5 poly
- Nir1 Q5 poly
- Nir1 2%DMSO Q5 poly
- Nir1 5%DMSO Q5 poly
- Nir1 3M Betaine Q5 poly
- AMO USER primers
- AMO +DMSO USER primers
- AMO +Betaine USER primers
- Nir1 DMSO
- Nir1 Betaine
- 1kb ladder
Conclusion
Today's PCRs on AMO and Nir were successful when DMSO was added.
Lab 115
Main purpose
Run Experiment 2. E.coli control measurement.
Who was in the lab
Ariadni, Natalia
Procedure
Adjusting the temperature at 37<up>o</sup> and calibrating the probes as described in Appendix 5.
Following the protocol Experiment 2
Changing the steps :
19. Add 0.5 ml of nitrite solution and continue by adding 0.5 ml after 10 minutes.
We added 0.5 ml nitrite again and then we took 2 ml of sample, we added then 0.5 ml and afterwards when there was any change in the curve we spiked with 0.4 ml of nitrite. We took 2 ml for sample in the end and we added 0.5 ml of N2O and 1 ml of NO to see if the probes are working properly.
Results
Colorimetric results
Measurements of standard solutions
Ammonium - 43.7 mg/L ( expected 39 mg/L)
Nitrite - < 2 mg/L (X10 diluted) -0.4 mg/L (with no dilution) (expected 0.5 mg/L)
Nitrate - 18.7 mg/L (X10 diluted) (expected 12 mg/L)
Measurement of samples
Ammonium
Measuring range 2-75 mg/L NH4-N
start point- no signal
middle point- signal <2 mg/L
end point- signal 0.7 mg/L
Nitrate
Measuring range 1-25 mg/L NO3-N
start point- no signal
middle point- signal <1 mg/L
end point- signal <1 mg/L
Nitrite
Measuring range 0.02-1 mg/L NO2-N
start point- no signal
middle point- signal 0.43 mg/L
end point- signal 0.79 mg/L
Conclusion
Problems with the NO probe and there were not proper results.
Navigate to the Previous or the Next Entry
 "
"