Team:UGent/Project
From 2013.igem.org
| Line 146: | Line 146: | ||
<html> | <html> | ||
| + | </html> | ||
| + | {{:Team:UGent/Templates/ToggleBoxStart}} Read why we chose <i>gfp</i> as gene of interest and T7 as inducible promoter {{:Team:UGent/Templates/ToggleBoxStart1}}{{:Team:UGent/Templates/ToggleBoxStart2}} | ||
| + | <html> | ||
| + | <h3>Gene of interest</h3> | ||
| + | <p>For our experiment, it would be useful to select a gene of interest that can easily be identified. Using a fluorescent reporter gene such as <i>gfp</i> is the obvious choice. When GFP is brought to expression in <i>E. coli</i> this results in the production of green light when illuminated with an ultraviolet source. A great advantage of GFP is the fact that no substrate is needed for its expression. Hence, GFP is ideal to measure gene expression, as it is not limited by substrate availability. The intensity of the fluorescence is proportional to the GFP presence in the cell, providing a first indication of the number of <i>gfp</i> genes present in the cell. Consequently, fluorescence intensity could give us a first insight in the number of construct copies attained through chromosomal evolution.</p> | ||
| + | <h3>Inducible promoter for <i>ccdB</i></h3> | ||
| + | <p>To increase the gene copy number in the chromosome, we have to be able to regulate the amount of CcdB in the cell. To achieve this, an inducible promoter is used. An inducible promoter is a combination of an operator and a promoter. The operator may be switched on or off which controls the transcription of the gene. Sometimes it is possible to regulate transcription according to the amount of stimulus the promoter receives. </p> | ||
| + | <p>The <b>T7 promoter</b> is not recognised by the <i>E. coli</i> RNA polymerase. This allows for good regulation of the transcription of genes, because there is no transcription as long as there is no T7 RNAP present. By using an inducible promoter to regulate the production of T7 RNAP, it is possible to change the amount of transcription of the desired protein. This is usually done by an IPTG inducible promoter.</p> | ||
| + | <p>Other options we considered were the <b>arabinose promoter</b> and the <b>lactose promoter</b>. We chose the T7 promoter because it shows lower leaky expression than these two. Leaky expression is transcription that occurs when no stimulus is given to the promoter. For the transcription of a toxic protein as in our case, leaky expression has to be avoided because even a small amount of transcription can be a burden to the cell and restrict cell growth.</p> | ||
| + | <p>The T7 promoter can be induced by IPTG. By using different concentrations we try to find a correlation between the applied IPTG concentration and the amount of gene copy numbers in the genome.</p> | ||
| + | </html> | ||
| + | {{:Team:UGent/Templates/ToggleBoxEnd}} | ||
| + | <html> | ||
<h1>Literature study</h1> | <h1>Literature study</h1> | ||
Revision as of 15:27, 14 September 2013
|
You can read our entire literature study about our new model for stabilised gene duplication here. Below you can read the most important aspects of it. Introduction: high gene expression in industrial biotechnologyThe main goal of industrial biotechnology is to increase the yield of biochemical products using microorganisms as production hosts. This includes engineering large synthetic pathways and improving their expression. Overexpression of genes has mainly been achieved by using high or medium copy plasmids. However, studies have demonstrated that plasmid-bearing cells lose their productivity fairly quickly as a result of genetic instability.
In industrial biotechnology, a common technique to express new synthetic products and pathways is the use of plasmids as vectors. Plasmids are easy to insert into cells and replicate independently from the genome, allowing strong gene expression. Overexpression is easily achieved by using plasmids with a medium or high copy number, different promoter systems, ribosome binding sites (RBS), etc. Thanks to plasmids, the industrial biotechnology has grown substantially over the past years. However, the use of plasmids entails some important disadvantages: plasmid maintenance imposes a metabolic burden on cells and plasmids suffer from genetic instability. Metabolic burdenWhen plasmids are present in cells and replicate, they create a metabolic burden. This is defined by Bentley et al. (1990) as “the amount of resources that are taken from the host cell metabolism for foreign DNA maintenance and replication”. As a result, metabolic load causes many alterations to the physiology and metabolism of the cell and reduces the cellular fitness. The most common change is a delayed growth. This can be caused by the fact that new pathways for energy generation are activated due to the competition between cell propagation and plasmid replication. This growth retardation leads to a lower yield of the desired product. Genetic instabilityPlasmids are genetically instable due to three processes: segregational instability, structural instability and allele segregation.
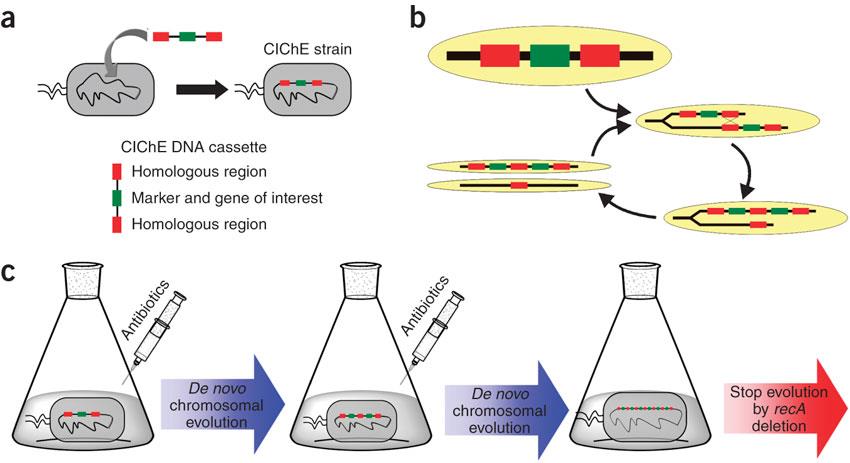
Therefore a new method was developed for the overexpression of a gene of interest in the bacterial chromosome: Chemically Inducible Chromosomal evolution (CIChE). In this technique the chromosome is evolved to contain a higher number of gene copies by adding a chemical inducer.
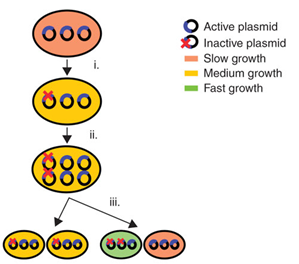
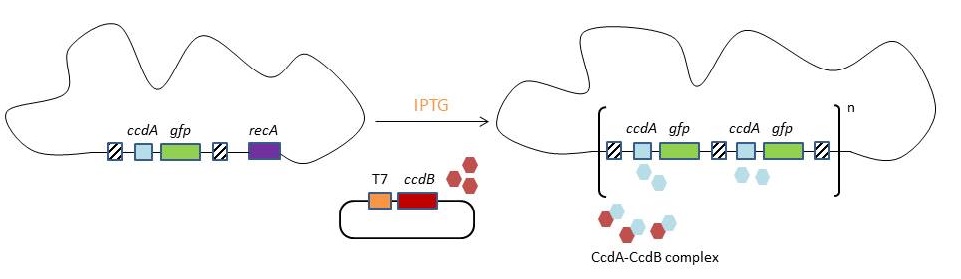
In 2009, Tyo et al. developed a technique for the stable, high copy expression of a gene of interest in E. coli without the use of high copy number plasmids, thus avoiding their previously stated negative characteristics. They called this plasmid-free, high gene copy expression system ‘chemically inducible chromosomal evolution’. In this method, the gene of interest is integrated in the microbial genome and then amplified to achieve multiple copies and reach the desired expression level. Genomic integration guarantees ordered inheritance, resolving the problem of allele segregation. CIChE works as follows: First a construct, containing the gene(s) of interest and the antibiotic marker chloramphenicol acetyl transferase (cat) flanked by homologous regions, is delivered to and subsequently integrated into the E. coli genome. The construct can be amplified in the genome through tandem gene duplication by recA homologous recombination. Then the strain is cultured in increasing concentrations of chloramphenicol, providing a growth advantage for cells with increased repeats of the construct and thereby selecting for bacteria with a higher gene copy number (Figure).
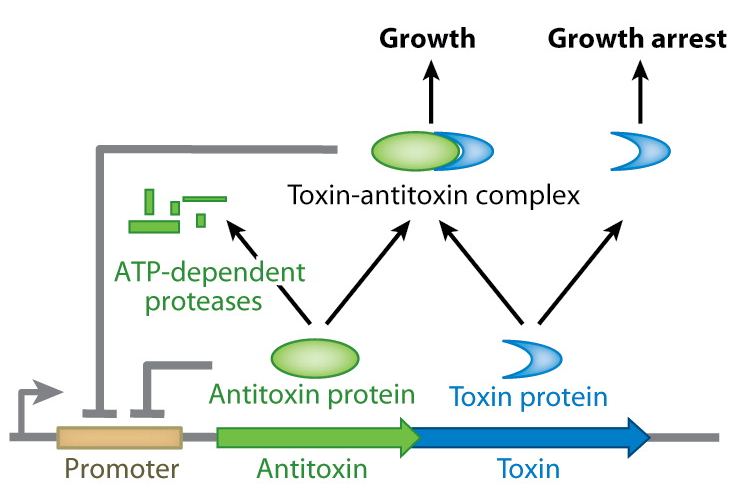
This process is called chromosomal evolution. When the desired gene copy number is reached recA is deleted, thereby fixing the copy number. The use of CIChE-strains poses several advantages. They require no selection markers and can be cultured without the addition of antibiotics to the medium. This is in contrast to strains containing plasmids, which still need antibiotic selection. Also, in CIChE-strains yields can be tuned by varying the antibiotic concentration during chromosomal evolution. CloseIt has been shown that approximately 40 and even up to 50 gene copies can be attained using chromosomal evolution. It has also been demonstrated that, while plasmid-bearing strains lose their productivity after 40 generations due to allele segregation, gene copy number and productivity of CIChE-strains remain stable even after 70 generations. This genetic stability is considered to be the most important asset of CIChE. The original model for CIChE, however, results in bacterial strains containing a large number of antibiotic resistance genes. To make this valuable technique more widely applicable in the industry, we developed a model for chromosomal evolution based on a toxin-antitoxin system instead of antibiotic resistance. | |||
 "
"