Team:HIT-Harbin/Experiments
From 2013.igem.org
| Line 86: | Line 86: | ||
<p>Table 2 Dilution of Two groups of bacteria</p> | <p>Table 2 Dilution of Two groups of bacteria</p> | ||
<img src="https://static.igem.org/mediawiki/2013/2/21/Experiment7.png"/> | <img src="https://static.igem.org/mediawiki/2013/2/21/Experiment7.png"/> | ||
| + | <img src="https://static.igem.org/mediawiki/2013/7/74/Experiment9.png"/> | ||
| + | <p>linear regression for absorbance vs relative concentration of RFP</p> | ||
</div> | </div> | ||
Revision as of 06:11, 27 September 2013
Project/experiments
Experiment Schedule
JuL. 10~15 Preparation of media, competent cells and experimental reagents JuL. 16~20 Preparation of parts from iGEM JuL. 21~31 Respective ligation of strong, intermediate and weak RBS with sub-circuit hrpR/hrpS/tet/RFP Aug. 1~10 Ligation of terminators Aug. 11~20 Successful ligation of the four sub-circuits Aug. 21~28 Combination of sub-circuits: the device Aug. 29~Sep. 15 Test of the device Sep. 16~25 Remaining experimentsLigation for our device
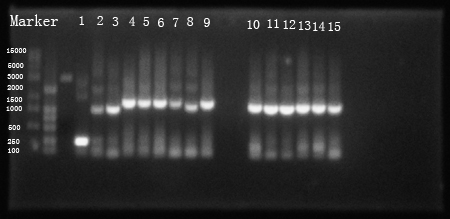
Fig 1. PCR resuLts of our own parts
(1: hrpL; 2: hrpS; 3: hrpR; 4: Ptet +strong RBS+hrpS+T; 5:Ptet+intermediate RBS+hrpS+T; 6: Ptet+weak RBS+hrpS+T; 7: PIPTG +strong RBS+hrpR+T; 8 is not needed; 9: PIPTG +weak RBS+hrpR+T; 10: PhrpL+strong RBS+tetR+T; 11: PhrpL+intermediate RBS+tetR+T; 12: PhrpL+weak RBS+tetR+T; 13: PhrpL+strong RBS+RFP+T; 14: PhrpL+weak RBS+RFP+T; 15: PhrpL+weak RBS+RFP+T)

Fig 1.EcoR1 and Pst1 double restriction enzyme cleavage for hrpL AND gate(BBa_K1014014) device and B-POM1(BBa_K1014999)
1:plasmid carrying BBa_K1014014; 2:double restriction enzyme cleavage for 1; 3:plasmid carrying BBa_K1014999; 4:double restriction enzyme cleavage for 3
1.Test of device
1)Preparation of IPTG solution: add 240mg IPTG powder into 10mL dd H2O. We filtrated the solution to sterilize it and broke it into EP tubes. The concentration is 24mg/mL (100mM/mL). then we stored them in -20℃. 2)To test the four different combination according to the strength of promoters, we made different concentrations of IPTG for the bacteria.Table 1 IPTG formula


Fig 3. No red after night
 Fig 4. Constitutive promoter expressing RFP
(Left is 12h, right is 24h)
Fig 4. Constitutive promoter expressing RFP
(Left is 12h, right is 24h)
2.Investigating the relationship between the concentration of RFP with that of IPTG
1)Measuring absorbance of RFP We grew bacteria without device and bacteria with our device in same volume until stationary phase. Taking bacteria without device as background, we measured the absorbance of bacteria with our device (the max absorption peak is 504nm). Before the mensuration, we diluted the two groups according to table2. We took the mean of two measures as the useful data.
Fig.5 RFP absorbance varying with wave length
Table 2 Dilution of Two groups of bacteria


linear regression for absorbance vs relative concentration of RFP
 "
"
