Team:Imperial College/Modelling beta-amyloid
From 2013.igem.org
| (23 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<h2>Introduction</h2> | <h2>Introduction</h2> | ||
| - | <p align="justify"> This is another degradation model we designed to help UCL 2013 iGEM team with their [https://2013.igem.org/Team:UCL/Project/Degradation Beta-Amyloid Degradation Assays]. Beta-Amyloid is known as the component of Amyloids Plaque in association with | + | <p align="justify"> This is another degradation model we designed to help UCL 2013 iGEM team with their [https://2013.igem.org/Team:UCL/Project/Degradation Beta-Amyloid Degradation Assays]. Beta-Amyloid is known as the component of Amyloids Plaque in association with [https://2013.igem.org/Team:UCL/Background/Alzheimers Alzheimer's disease (AD)], in which the target of UCL iGEM this year. Matrix metallopeptidase 9 (MMP-9) was discovered to be able to degrade Beta-Amyloid that can be potentially used to cure Alzheimer's disease. The model provides simulations about how efficient is MMP-9 enzyme to degrade Beta-Amyloids.</p> |
<h2>Description</h2> | <h2>Description</h2> | ||
<h3>Objective</h3> | <h3>Objective</h3> | ||
| - | <p align="justify"> The Mathematical model | + | <p align="justify"> The Mathematical model was built in MATLAB extension Simbiology, it will predict how long will take to degrade a certain concentration of Beta-Amyloid by a given amount of MMP-9 enzyme. The simulation results can help UCL iGEM team to modify their assays in order to achieve better experimental results. The model can also be used to provide a preliminary prediction of degrading Amyloid Plaque in vivo.</p> |
<h3>Degradation Assays of UCL 2013 iGEM team</h3> | <h3>Degradation Assays of UCL 2013 iGEM team</h3> | ||
| + | The details of the assays are [https://2013.igem.org/Team:UCL/Project/Degradation here]. There are three different substrates which are soluble aggregated β-amyloid (Aβ),aggregated β-amyloid (Aβ) fibrils 1-40 and aggregated β-amyloid (Aβ) fibrils 1-42. Here is the table that defines the initial concentrations of all substrates and the enzyme from the assays. | ||
| + | <table border="2" bgcolor="#efefef"> | ||
| + | <tr> | ||
| + | <th width="150">Species</th> | ||
| + | <th width="150">Concentration</th> | ||
| + | <th width="150">Units</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>soluble Aβ</th> | ||
| + | <td>25</td> | ||
| + | <td>uM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Aβ fibrils 1-40</th> | ||
| + | <td>200</td> | ||
| + | <td>uM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>Aβ fibrils 1-42</th> | ||
| + | <td>100</td> | ||
| + | <td>uM</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <th>MMP-9</th> | ||
| + | <td>0.2</td> | ||
| + | <td>uM</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | <h2>Methods</h2> | ||
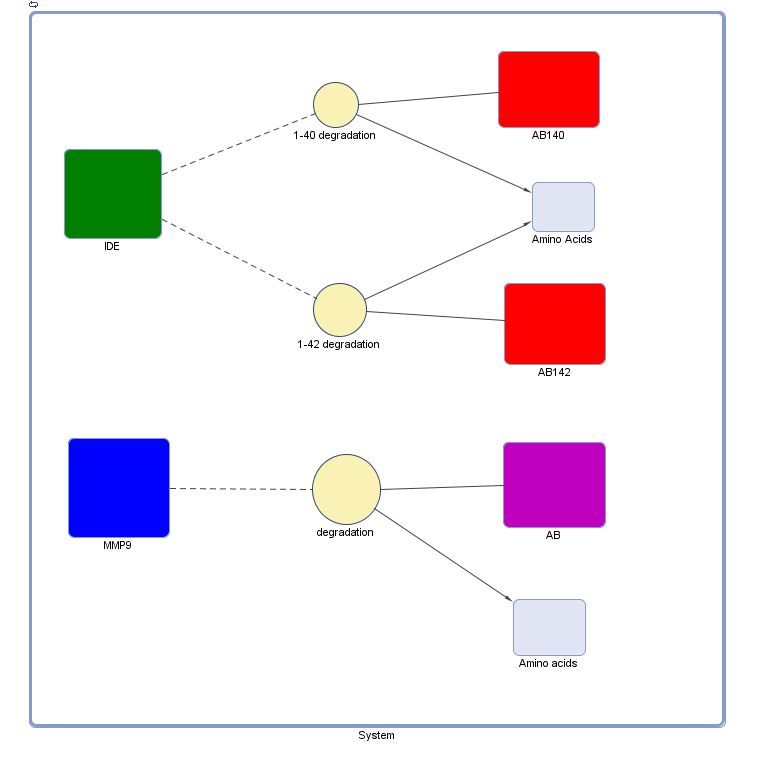
| + | <h3>The Model</h3> | ||
| + | [[File:BAmyloidModel.png]] | ||
| + | <p>Explanation</p> | ||
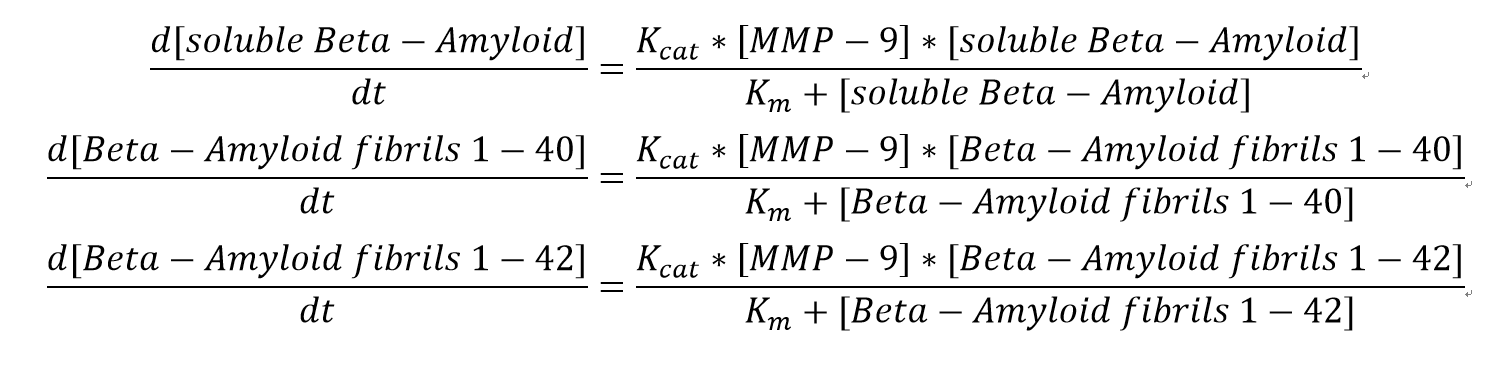
| + | <h3>Equations</h3> | ||
| + | <p align="justify">We choose simple Michalis-Menten mechanism for Beta-Amyloid degradation. The ODEs for all substrate are:</p> | ||
| + | [[File:Simple MM.png]] | ||
| + | <p>The Km values here are for each substrate correspondingly whereas Kcat values are for MMP-9 in each assay. </p> | ||
| + | <h3>Assumption</h3> | ||
| + | <p align="justify"> Although Matrix metallopeptidase 9 (MMP-9) is a well characterised enzyme, there is no clear kinetic data for degrading soluble Beta-Amyloids and its fibrils. | ||
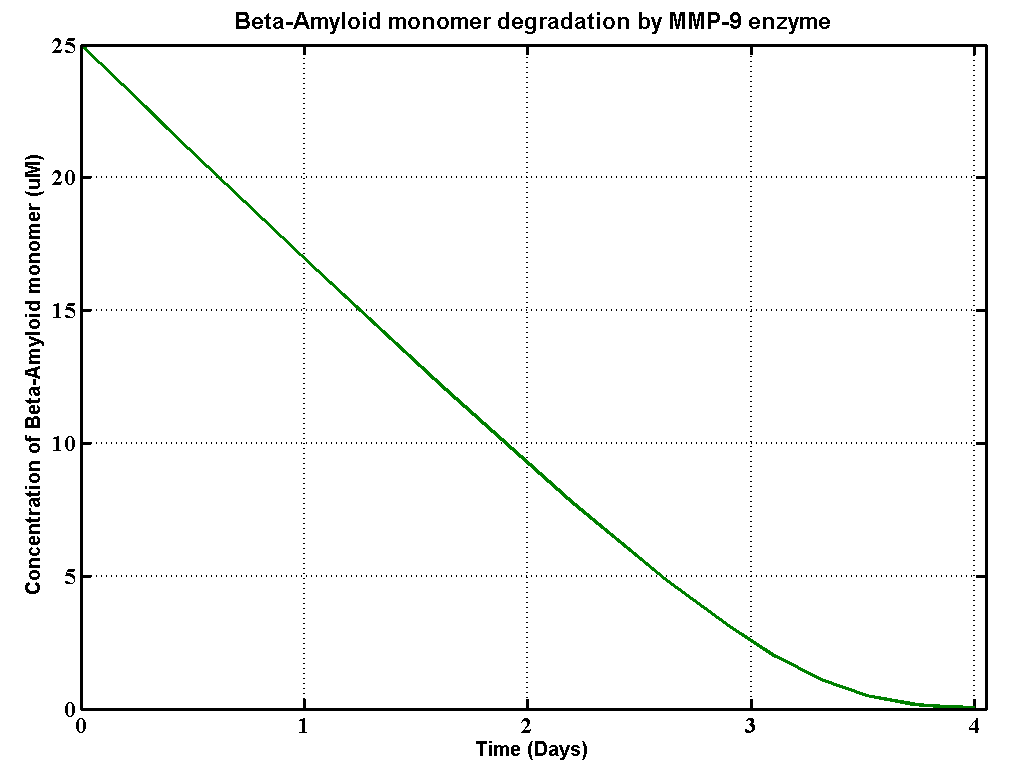
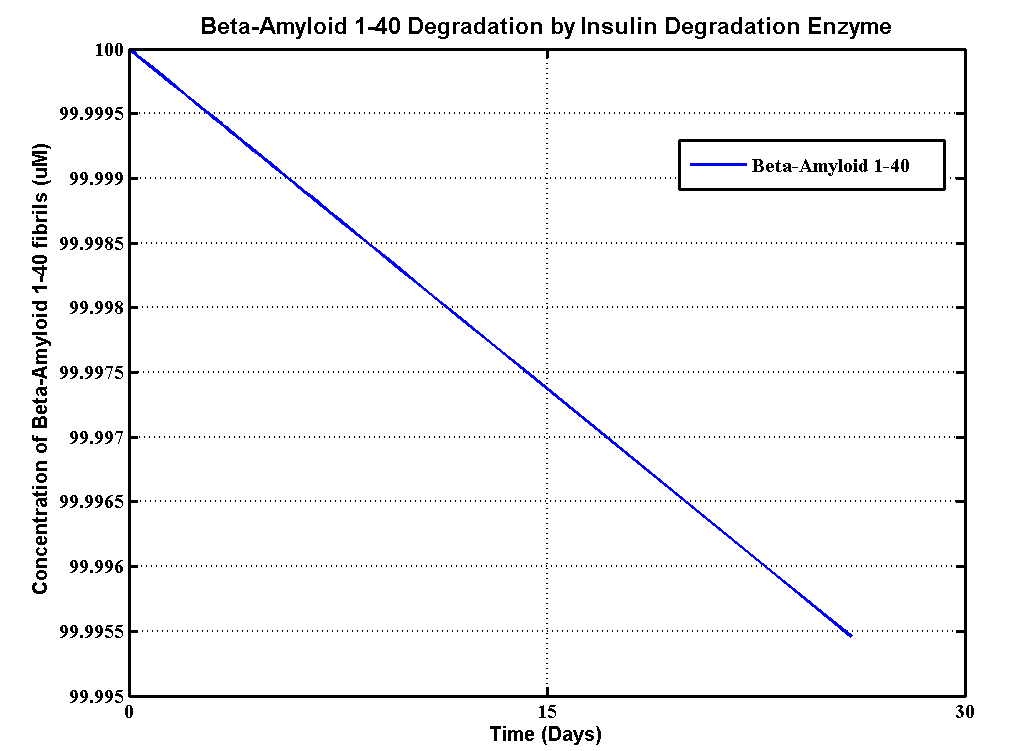
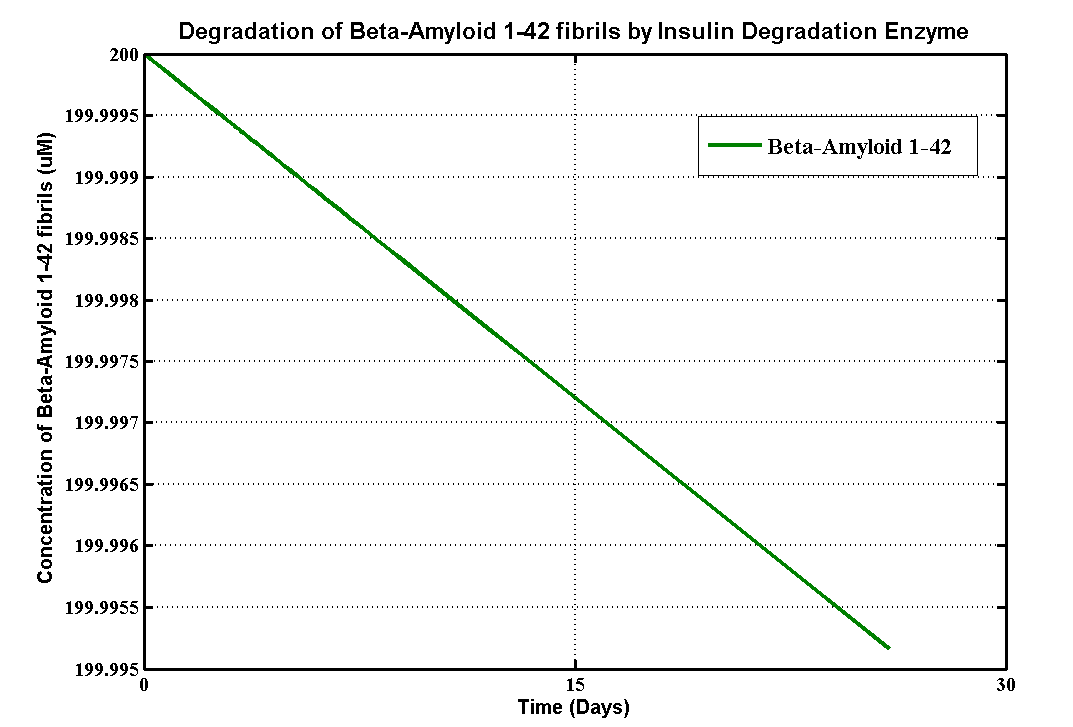
| + | <h2>Results</h2> | ||
| + | [[File:Mmp9.png]] | ||
| + | [[File:BA140.png]] | ||
| + | [[File:BA142.png]] | ||
| + | <h2>Reference</h2> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| - | |||
{{:Team:Imperial_College/Templates:footer}} | {{:Team:Imperial_College/Templates:footer}} | ||
Latest revision as of 19:20, 1 October 2013
Contents |
Beta-Amyloid Degradation Model
Introduction
This is another degradation model we designed to help UCL 2013 iGEM team with their Beta-Amyloid Degradation Assays. Beta-Amyloid is known as the component of Amyloids Plaque in association with Alzheimer's disease (AD), in which the target of UCL iGEM this year. Matrix metallopeptidase 9 (MMP-9) was discovered to be able to degrade Beta-Amyloid that can be potentially used to cure Alzheimer's disease. The model provides simulations about how efficient is MMP-9 enzyme to degrade Beta-Amyloids.
Description
Objective
The Mathematical model was built in MATLAB extension Simbiology, it will predict how long will take to degrade a certain concentration of Beta-Amyloid by a given amount of MMP-9 enzyme. The simulation results can help UCL iGEM team to modify their assays in order to achieve better experimental results. The model can also be used to provide a preliminary prediction of degrading Amyloid Plaque in vivo.
Degradation Assays of UCL 2013 iGEM team
The details of the assays are here. There are three different substrates which are soluble aggregated β-amyloid (Aβ),aggregated β-amyloid (Aβ) fibrils 1-40 and aggregated β-amyloid (Aβ) fibrils 1-42. Here is the table that defines the initial concentrations of all substrates and the enzyme from the assays.
| Species | Concentration | Units |
|---|---|---|
| soluble Aβ | 25 | uM |
| Aβ fibrils 1-40 | 200 | uM |
| Aβ fibrils 1-42 | 100 | uM |
| MMP-9 | 0.2 | uM |
Methods
The Model
Explanation
Equations
We choose simple Michalis-Menten mechanism for Beta-Amyloid degradation. The ODEs for all substrate are:
The Km values here are for each substrate correspondingly whereas Kcat values are for MMP-9 in each assay.
Assumption
Although Matrix metallopeptidase 9 (MMP-9) is a well characterised enzyme, there is no clear kinetic data for degrading soluble Beta-Amyloids and its fibrils.
Results
Reference
 "
"