Team:Edinburgh/Introduction/Aggregation
From 2013.igem.org
Hristianita (Talk | contribs) |
|||
| Line 3: | Line 3: | ||
<div class='content'> | <div class='content'> | ||
| + | |||
| + | '''<b>Aggregation</b>''' | ||
| + | |||
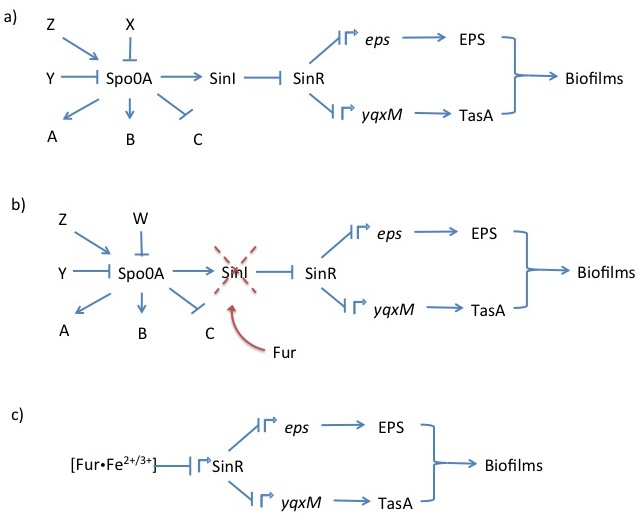
| + | <i>Bacillus subtilis</i> naturally forms biofilms composed of exopolysaccharides and the protein TasA. Production of both of these components is repressed by the transcription factor SinR. It represses the <i>eps</i> (exopolysaccharide) and <i>yqxM</i> (includes TasA) operons by binding DNA at their regulatory elements. The <i>eps</i> and <i>yqxM</i> operons contain various genes encoding biofilm formation enzymes and regulatory elements. The repressive functionality of SinR is disrupted by SinI, which binds to SinR disables its’ ability to attach to DNA (Whole pathway illustrated in Figure 1a). <sup>1</sup> The upstream regulation of SinI and SinR is very complex and the details are not important for this project. | ||
| + | |||
| + | <b>Novelty in this project</b> | ||
| + | |||
| + | Upon forming a biofilm, the cells aggregate into a floating pellicle if grown in liquid. In our project, we want to use this natural <i>B. subtilis</i> feature as a method of extraction. Furthermore, we intend to make biofilm formation responsive to iron ion concentration in the environment recognisable by the Fur transcription factor. We will attempt to accomplish this by replacing the SinI gene, which is directly downstream of SinR, and normal SinR regulatory elements by a constitutive promoter with a Fur binding box. This would cause biofilm formation derepression in the presence of iron (Figure 1b & 1c). | ||
| + | |||
| + | <i><b>Figure 1</b></i> Schematic showing the most crucial regulatory sequence elements from the <i>B. subtilis</i> master regulator Spo0A, which integrates various cellular signals, to operons and their gene products that create biofilms (a). The genetic modifications we intend to perform in our aggregation subproject (b) (c). <sup>1</sup> | ||
| + | [[Image:EdiAggregation.jpg|600px|center]] | ||
| + | |||
</div> | </div> | ||
{{Team:Edinburgh/Footer}} | {{Team:Edinburgh/Footer}} | ||
Revision as of 17:14, 3 October 2013
Aggregation
Bacillus subtilis naturally forms biofilms composed of exopolysaccharides and the protein TasA. Production of both of these components is repressed by the transcription factor SinR. It represses the eps (exopolysaccharide) and yqxM (includes TasA) operons by binding DNA at their regulatory elements. The eps and yqxM operons contain various genes encoding biofilm formation enzymes and regulatory elements. The repressive functionality of SinR is disrupted by SinI, which binds to SinR disables its’ ability to attach to DNA (Whole pathway illustrated in Figure 1a). 1 The upstream regulation of SinI and SinR is very complex and the details are not important for this project.
Novelty in this project
Upon forming a biofilm, the cells aggregate into a floating pellicle if grown in liquid. In our project, we want to use this natural B. subtilis feature as a method of extraction. Furthermore, we intend to make biofilm formation responsive to iron ion concentration in the environment recognisable by the Fur transcription factor. We will attempt to accomplish this by replacing the SinI gene, which is directly downstream of SinR, and normal SinR regulatory elements by a constitutive promoter with a Fur binding box. This would cause biofilm formation derepression in the presence of iron (Figure 1b & 1c).
Figure 1 Schematic showing the most crucial regulatory sequence elements from the B. subtilis master regulator Spo0A, which integrates various cellular signals, to operons and their gene products that create biofilms (a). The genetic modifications we intend to perform in our aggregation subproject (b) (c). 1

| 
| | | | 
|
| This iGEM team has been funded by the MSD Scottish Life Sciences Fund. The opinions expressed by this iGEM team are those of the team members and do not necessarily represent those of MSD | |||||
 "
"