Team:Edinburgh/Project/Results/Chassis
From 2013.igem.org
Krothschild (Talk | contribs) |
|||
| (15 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<div class='content'> | <div class='content'> | ||
| - | In order to better | + | <h3>Characterisation of ''Bacillus subtilis 168'' as a chassis</h3> |
| + | |||
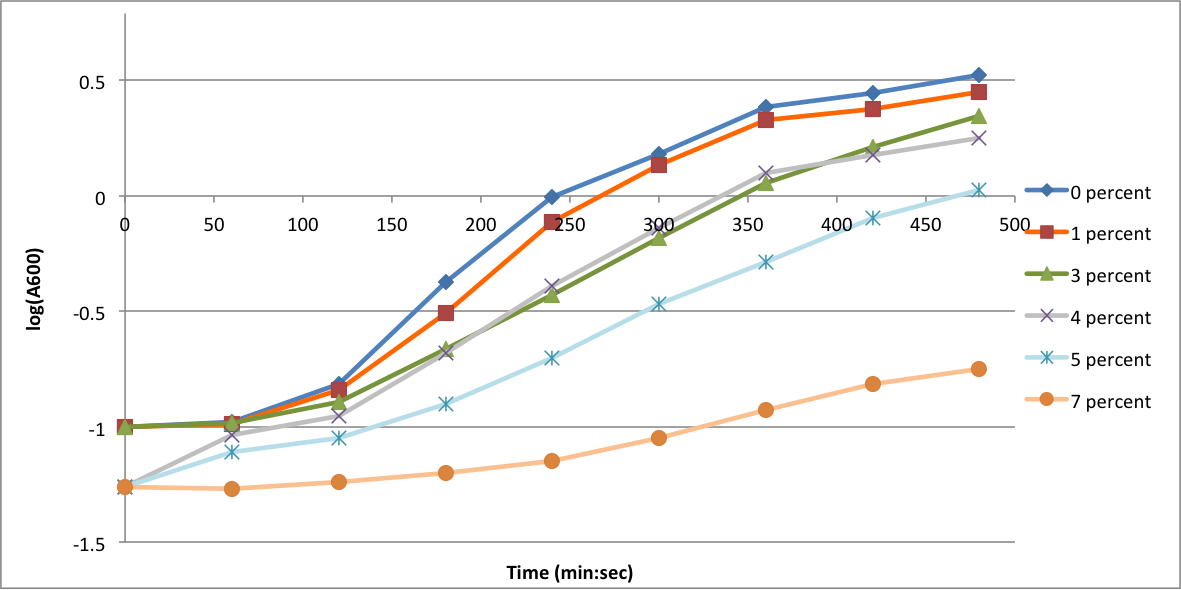
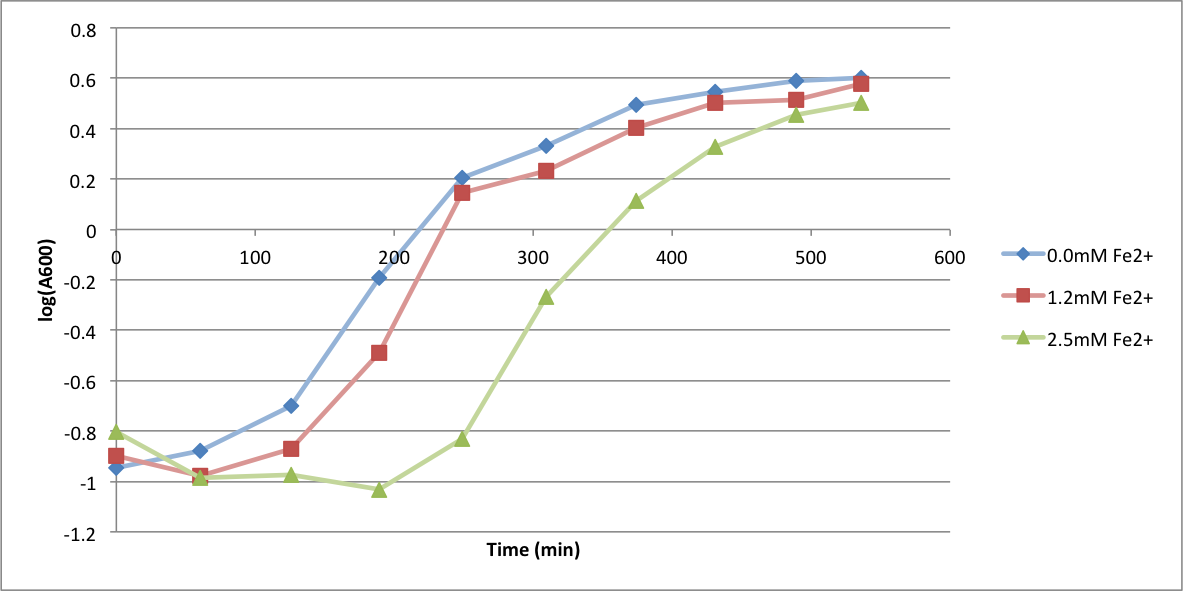
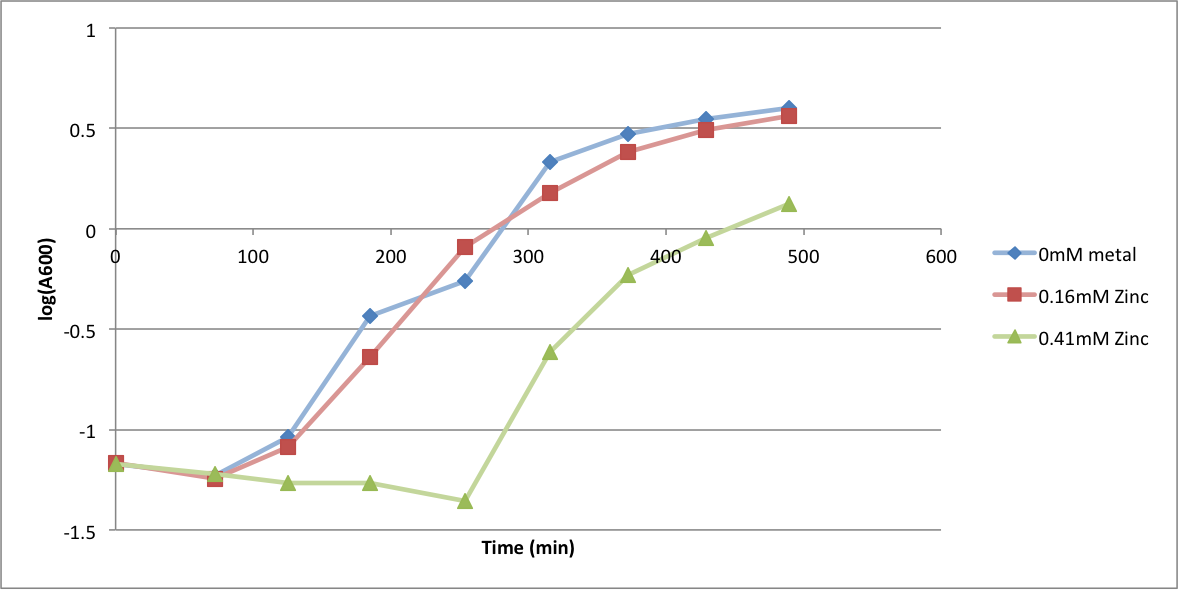
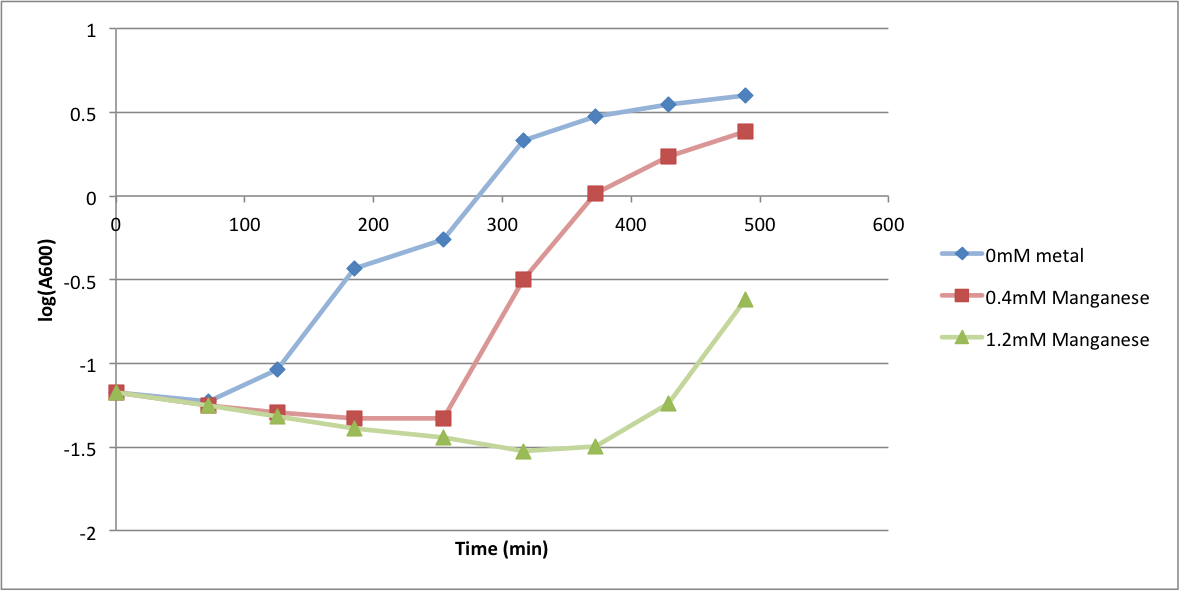
| + | In order to better understand what conditions are best suitable for growth of our experimental strain, <i> Bacillus subtilis</i> 168, we tested the growth under various conditions relevant to the work we would perform later. First we measured the growth curves at various ethanol concentrations to confirm the highest ethanol concentration suitable for cell growth, which was 4%, as displayed below. Afterwards, we measured the growth curves at various concentrations of iron, copper, nickel, cobalt, zinc and manganese, which are also displayed below. | ||
[[Image:EdiChassis.png|600px|center]] | [[Image:EdiChassis.png|600px|center]] | ||
[[Image:EdiChassis1.png|600px|center]] | [[Image:EdiChassis1.png|600px|center]] | ||
| - | |||
[[Image:EdiChassis2.png|600px|center]] | [[Image:EdiChassis2.png|600px|center]] | ||
[[Image:EdiChassis3.png|600px|center]] | [[Image:EdiChassis3.png|600px|center]] | ||
| Line 18: | Line 19: | ||
| - | <h3>Testing the Kanamycin resistance conferred by pTG262 (BBa_I742123) in B.subtilis and E.coli</h3> | + | <div id="vector"><h3>Testing the Kanamycin resistance conferred by pTG262 (BBa_I742123) in <i>B.subtilis</i> and <i>E.coli</i></h3></div> |
| - | B.subtilis 168 and E. coli JM109 were each transformed with the vector pTG262 (BBa_I742123) with an RFP biobrick insert cassette. | + | <i>B.subtilis</i> 168 and <i>E. coli</i> JM109 were each transformed with the vector pTG262 (BBa_I742123) with an RFP biobrick insert cassette. |
| - | <h2>B.subtilis</h2> | + | <h2><i>B.subtilis</i></h2> |
| - | + | Wild type ''B. subtilis'' 168 and pTG262 transformed ''B.subtilis'' 168 colonies were used to inoculate 5 ml of LB medium intp glass standard bottles which were incubated at 37° C with shaking for ~6h (pTG262 with 5 mg.L-1 chloramphenicol to prevent vector loss). This culture was then diluted by a factor of 10:6 and 100ul aliquots were plated onto LB agar plates of varying kanamycin concentrations, and subsequently incubated overnight at 37° C. | |
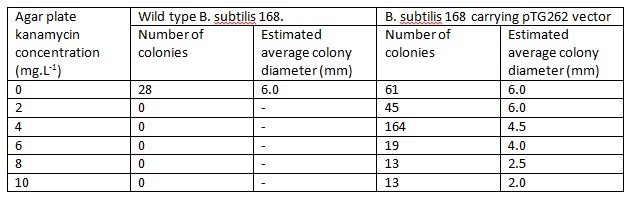
| - | The number and size of colonies which grew on each of the agar plates was recorded. | + | The number and size of colonies which grew on each of the agar plates was recorded as seen below. |
[[File:B.subAgar.jpg]] | [[File:B.subAgar.jpg]] | ||
| - | <h2>E.coli test of growth on agar plates</h2> | + | <h2><i>E.coli</i> test of growth on agar plates</h2> |
| - | + | Wild type <i>E.coli</i> JM109 and pTG262 transformed <i>E.coli</i> JM109 colonies were used to inoculate 5 ml LB medium in glass standard bottles which were incubated at 37° C with shaking for 6 hours and 30 minutes (pTG262 inoculated in duplicate with no antibiotic and 10 mg.L-1 Kanamycin). | |
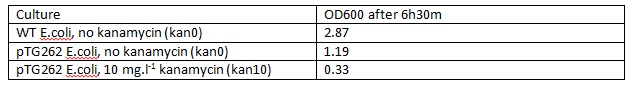
| - | The OD600 of the cultures was measured: | + | The OD600 of the cultures was measured as shown below: |
[[File:E.coliInoculumforplates.jpg]] | [[File:E.coliInoculumforplates.jpg]] | ||
| - | Each of the above cultures were diluted by factors of 10 | + | Each of the above cultures were diluted by factors of 10:5 and 10:6. Agar plates of varying kanamycin concentration were plated with 100 μl aliquots of the dilutions, and incubated for ~36 hr at 37° C (Figure 1). The number and colour of the colonies were recorded (red indicating presence of pTG262, white being wild type or representing loss of the plasmid before plating). |
[[File:E.coliAgar.jpg]] | [[File:E.coliAgar.jpg]] | ||
| Line 44: | Line 45: | ||
[[File:E.coliAgarpicture.jpg|600px|]] | [[File:E.coliAgarpicture.jpg|600px|]] | ||
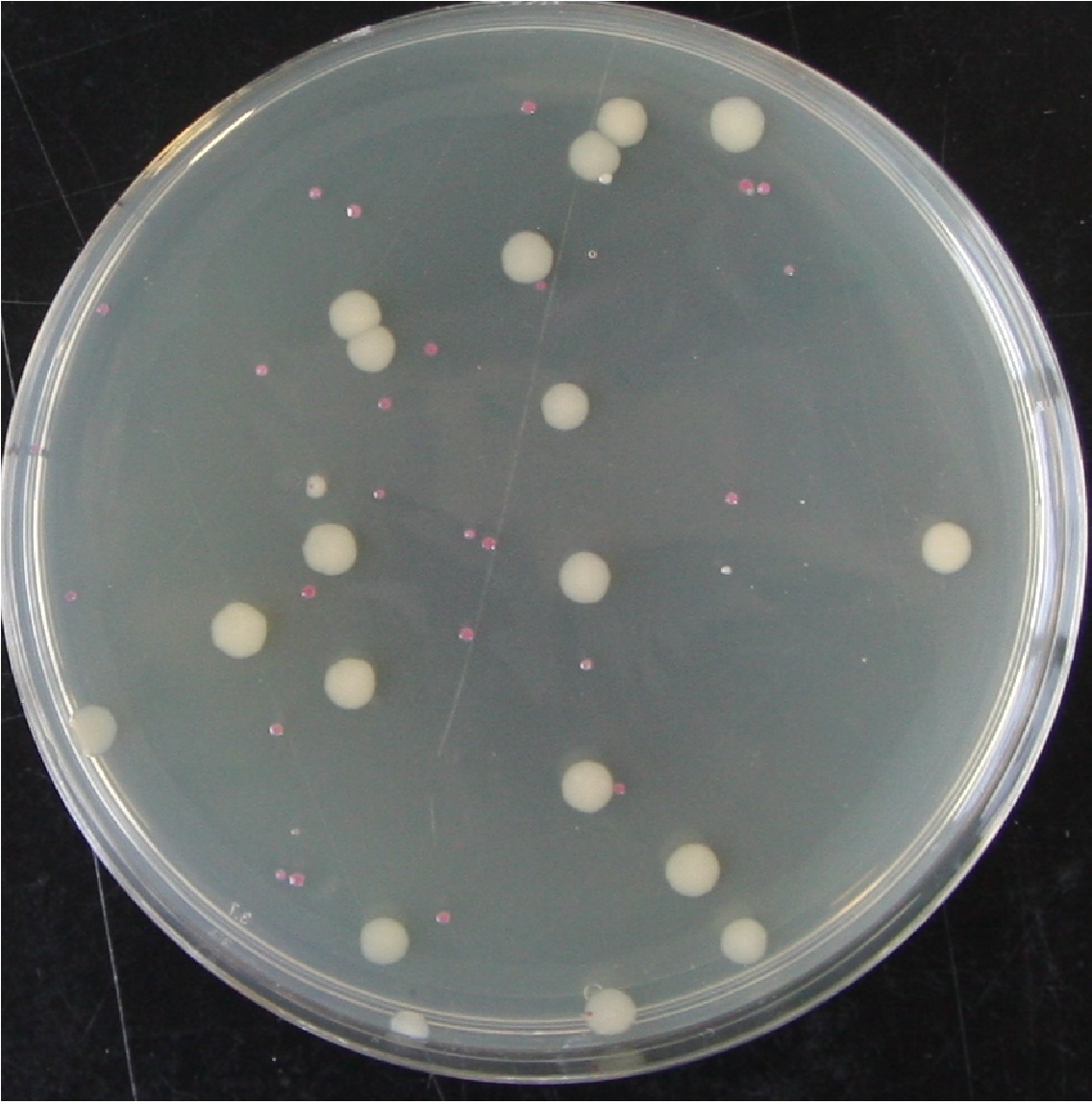
| - | Figure 1. A plate from the above experiment after an additional 24 | + | Figure 1. A plate from the above experiment after an additional 24 hr on the bench, pTG262 Kan0 10:5 dilution plated onto 0 mg.L-1 Kanamycin. This shows the typical colony size as indicated in the above table. It demonstrates both presumed plasmid loss in overnight culture without antibiotic (white colonies) and the decreased growth rate of cells containing pTG262. |
| - | <h2>E. coli test of growth in Kanamycin liquid culture</h2> | + | <h2><i>E. coli</i> test of growth in Kanamycin liquid culture</h2> |
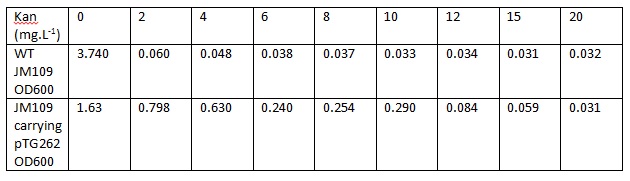
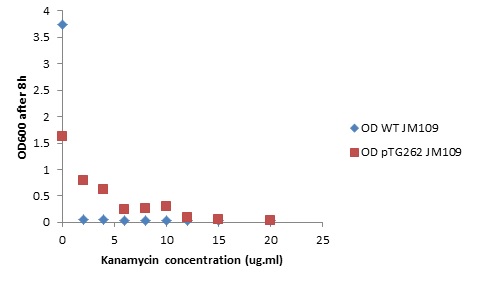
| - | Overnight LB liquid cultures of | + | Overnight LB liquid cultures of wild type <i>E.coli</i> JM109 and <i>E.coli</i> JM109 transformed with pTG262 were diluted 1:50 in fresh LB medium. 3 ml aliquots of each were put in glass standard bottles, along with varying amounts of kanamycin. The Optical density at 600 nm was measured at the start and after 8 hr incubation with shaking at 37° C. |
Initial OD600: WT JM109 - 0.030, pTG262 - 0.028. | Initial OD600: WT JM109 - 0.030, pTG262 - 0.028. | ||
| - | OD600 after 8 | + | OD600 after 8 hr incubation with shaking at 37° C: |
[[File:LiquidcultureE.coli.jpg]] | [[File:LiquidcultureE.coli.jpg]] | ||
Latest revision as of 23:14, 4 October 2013
Contents |
Characterisation of Bacillus subtilis 168 as a chassis
In order to better understand what conditions are best suitable for growth of our experimental strain, Bacillus subtilis 168, we tested the growth under various conditions relevant to the work we would perform later. First we measured the growth curves at various ethanol concentrations to confirm the highest ethanol concentration suitable for cell growth, which was 4%, as displayed below. Afterwards, we measured the growth curves at various concentrations of iron, copper, nickel, cobalt, zinc and manganese, which are also displayed below.
Testing the Kanamycin resistance conferred by pTG262 (BBa_I742123) in B.subtilis and E.coli
B.subtilis 168 and E. coli JM109 were each transformed with the vector pTG262 (BBa_I742123) with an RFP biobrick insert cassette.
B.subtilis
Wild type B. subtilis 168 and pTG262 transformed B.subtilis 168 colonies were used to inoculate 5 ml of LB medium intp glass standard bottles which were incubated at 37° C with shaking for ~6h (pTG262 with 5 mg.L-1 chloramphenicol to prevent vector loss). This culture was then diluted by a factor of 10:6 and 100ul aliquots were plated onto LB agar plates of varying kanamycin concentrations, and subsequently incubated overnight at 37° C.
The number and size of colonies which grew on each of the agar plates was recorded as seen below.
E.coli test of growth on agar plates
Wild type E.coli JM109 and pTG262 transformed E.coli JM109 colonies were used to inoculate 5 ml LB medium in glass standard bottles which were incubated at 37° C with shaking for 6 hours and 30 minutes (pTG262 inoculated in duplicate with no antibiotic and 10 mg.L-1 Kanamycin).
The OD600 of the cultures was measured as shown below:
Each of the above cultures were diluted by factors of 10:5 and 10:6. Agar plates of varying kanamycin concentration were plated with 100 μl aliquots of the dilutions, and incubated for ~36 hr at 37° C (Figure 1). The number and colour of the colonies were recorded (red indicating presence of pTG262, white being wild type or representing loss of the plasmid before plating).
Figure 1. A plate from the above experiment after an additional 24 hr on the bench, pTG262 Kan0 10:5 dilution plated onto 0 mg.L-1 Kanamycin. This shows the typical colony size as indicated in the above table. It demonstrates both presumed plasmid loss in overnight culture without antibiotic (white colonies) and the decreased growth rate of cells containing pTG262.
E. coli test of growth in Kanamycin liquid culture
Overnight LB liquid cultures of wild type E.coli JM109 and E.coli JM109 transformed with pTG262 were diluted 1:50 in fresh LB medium. 3 ml aliquots of each were put in glass standard bottles, along with varying amounts of kanamycin. The Optical density at 600 nm was measured at the start and after 8 hr incubation with shaking at 37° C.
Initial OD600: WT JM109 - 0.030, pTG262 - 0.028.
OD600 after 8 hr incubation with shaking at 37° C:

| 
| | | | 
|
| This iGEM team has been funded by the MSD Scottish Life Sciences Fund. The opinions expressed by this iGEM team are those of the team members and do not necessarily represent those of MSD | |||||
 "
"