Team:Imperial College/Protocols
From 2013.igem.org
| (72 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{:Team:Imperial_College/Templates:header}} | |
| - | {{:Team:Imperial_College/Templates | + | |
| - | + | ||
| - | + | ||
<div id='maincontent'> | <div id='maincontent'> | ||
| Line 8: | Line 5: | ||
<h1>Materials</h1> | <h1>Materials</h1> | ||
| - | < | + | <h3>Waste Conditioned Media (WCM)</h3> |
| - | <p>We added 1g[http://en.wikipedia.org/wiki/Refuse-derived_fuel SRF] (Solid Recovered Fuel)/50 mL LB and then autoclaved the mixture. Once autoclaved, large waste chunks were removed through filter sterilisation (0. | + | <p>We added 1g [http://en.wikipedia.org/wiki/Refuse-derived_fuel SRF] (Solid Recovered Fuel) /50 mL LB and then autoclaved the mixture. Once autoclaved, large waste chunks were removed through filter sterilisation (0.2 µm filters) before being used in growth assays as waste conditioned media (WCM). |
| + | |||
| + | SRF refers to the refuse from recycling facilities that has no value currently and is incinerated to produce power at a cost to the recycling facility, it is composed of 30% plastics while the rest is cellulosic waste</p> | ||
| + | |||
| + | <h3>3% glucose Nile Red plates</h3> | ||
| + | <p>When making up LB agar media, add 3% glucose then autoclave. When autoclaving it is <b>essential</b> that you keep temperature <125°C. Failure to do so will result in caramelisation (160°C) of the media, which will show as a darkening of the media. With this complete, 60 µL Nile Red was added in addition to 150 µL chloramphenicol in a 300 mL final volume. Plates were then poured and left to dry. Another successful mechanism to thwart caramelisation is to autoclave the LB-agar then add the 3% glucose after. Follow this with a 30 second microwave step and then an 80°C water bath for a couple hours to thoroughly sterilise.</p> | ||
| + | |||
| + | <h3>M9 minimal and supplemented media</h3> | ||
| + | First made stocks of MgSO4 (246.48 g/mol), CaCl2 (147.02 g/mol), and thiamine hydrochloride (337.3 g/mol): | ||
| + | MgSO4: 100 mM - 2.46g in 100 mL ddH2O | ||
| + | CaCl2: 20 mM - 0.29g in 100 mL ddH2O | ||
| + | Thiamine hydrochloride: 50 mM - 0.84g in 50 mL ddH2O | ||
| + | The bottles used were pre-autoclaved. When all media was prepared, it was sterile filtrated, as autoclaving results in the formation of magnesium precipitate. See below for correct volumes and mass to add where [final] represents desired values. | ||
| + | <br> | ||
| + | [[File:M9_table.PNG]] | ||
| + | <br> | ||
| + | |||
| + | <h3>Xylose and Arabinose</h3> | ||
| + | For induction assays, 2% xylose was prepared, as was 6 µM arabinose. | ||
| + | |||
| + | |||
| - | |||
<h1>Protocols and Assays</h1> | <h1>Protocols and Assays</h1> | ||
| - | <br> <br> | + | https://static.igem.org/mediawiki/2013/c/c9/Mixed_waste2.jpg |
| - | <b>Waste | + | |
| + | |||
| + | <h3>P3HB production and extraction</h3> | ||
| + | |||
| + | <h4>Extracting PHB</h4> | ||
| + | <ul> | ||
| + | <li>Centrifuge settings: 4000RPM, 10mins.</li> | ||
| + | <li> Scale as appropriate.</li> | ||
| + | <li> After each centrifuge step the supernatant should be poured off. </li> | ||
| + | <li> This should provide PHB with 99% purity and a high molecular weight. </li> | ||
| + | </ul> | ||
| + | <ol> | ||
| + | <li>Centrifuge 50ml culture and resuspend in PBS.</li> | ||
| + | <li>Resuspend pellet in 5ml Triton X-100(1% v/v in PBS) for 30mins at room temp. </li> | ||
| + | <li>Centrifuge, resuspend in 5ml PBS.</li> | ||
| + | <li>Centrifuge, add 5ml sodium hyperchlorite solution and incubate at 30˚C for 1 hour.</li> | ||
| + | <li>Centrifuge, wash with 5ml distilled water and 70% EtOH several times.</li> | ||
| + | <li>Allow powder to dry.</li> | ||
| + | </ol> | ||
| + | |||
| + | <p>Shahryar Shakeri, Comparison of intracellular polyhydroxybutyrate granules formation between different bacterial cell subpopulations by flow cytometry, Jundishapur Journal of Microbiology 2011</p> | ||
| + | [http://jjmicrobiol.com/?page=article&article_id=2342] | ||
| + | |||
| + | <h4>LB-Agar Plates containing Nile Red stain</h4> | ||
| + | <p>Nile Red final concentration in media should be 0.5µg/ml</p> | ||
| + | |||
| + | <ol> | ||
| + | <li>After autoclaving 300ml of LB-Agar, add 600µl Nile Red solution(0.25mg/ml DMSO) </li> | ||
| + | <li>Add 150µl chloramphenicol </li> | ||
| + | <li>Pour plates in a clean fume cupboard</li> | ||
| + | </ol> | ||
| + | |||
| + | <h4>Nile Red Culture staining</h4> | ||
| + | <p>Nile Red final concentration should be 20µg/ml media</p> | ||
| + | |||
| + | <p>Add 320µl to 4 ml overnight culture and incubate for 30mins at room temp. </p> | ||
| + | |||
| + | <h4>Fluorescent Microscopy</h4> | ||
| + | <p>Agar pads are first made</p> | ||
| + | <ol> | ||
| + | <li>Small pads are made using 1% agarose in M9 minimal media between microscope slides. </li> | ||
| + | <li>2ul of the sample is added to each pad and imaged with the fluorescent microscope. </li> | ||
| + | </ol> | ||
| + | |||
| + | |||
| + | <h4>Relationship between OD and dry biomass for phaCAB operon containing cells</h4> | ||
| + | <ol> | ||
| + | <li>Grow 3 large cultures in LB and 3% glucose - inoculate with 1ml OD 0.1 and grow for 24h. </li> | ||
| + | <li>Calculate OD of original culture and dilute until there is a range of 0.2 - 1.0 OD(try to have large volumes for low OD). </li> | ||
| + | <li>Weigh centrifuge containers. </li> | ||
| + | <li>Transfer solutions to these containers, recording what volume of each OD is transferred. . </li> | ||
| + | <li>Centrifuge these solutions down and dry the pellet out until constant weight in a 50°C oven. </li> | ||
| + | <li>Dry cell mass = (container mass + dry cell mass)/container mass </li> | ||
| + | <li>Plot OD against dry cell mass. </li> | ||
| + | </ol> | ||
| + | |||
| + | <p></p> | ||
| + | |||
| + | <h2>Cell lysate activity assay</h2> | ||
| + | <ol> | ||
| + | <li><b>Preparation of the cell lysate:</b></li> | ||
| + | 5 mL cultures of EstC2 [http://parts.igem.org/Part:BBa_K1149002 BBa_K1149002], PueB [http://parts.igem.org/Part:BBa_K1149004 BBa_K1149004] and PulA [http://parts.igem.org/Part:BBa_K1149006 BBa_K1149006] were incubated overnight. The next morning, these were diluted in 200 mL and incubated until the OD reached 0.5-0.6. | ||
| + | The cultures were then induced with 6mM arabinose (final concentration)and left overnight. The following morning the cells were spun down, resuspended in 100 mM potassium phosphate buffer of pH 7.4 and probe-sonicated with 2 minute cycles of 2 seconds on, 2 seconds off, at 60% amplitude. The samples were kept on ice throughout the sonication process. The cell debris was pelleted by spinning and the supernatant was removed for utilising in the enzyme activity assays.</li> | ||
| + | <br><br><li><b>Substrate preparation:</b></li> | ||
| + | The substrate was prepared by the addition of 8.8 µL of para-Nitrophenyl butyrate to 1 mL of Acetonitril. | ||
| + | <br><li><b>Incubation of the reagents:</b></li> | ||
| + | In 10mm cuvettes, 5 µL of the EstC2, PueB, PulA, and Empty vector were added to 1 mL of 100 mM Potassium Phosphate buffer of pH 7.4 and incubated for 2-3 minutes. For the Substrate alone assay, simply 1 mL of buffer was incubated. | ||
| + | <br><li><b>Spectrometer programme:</b></li> | ||
| + | The Eppendorf BioSpectrometer was set up to run for 600 seconds, recording the absorbance at 405 nm every 30 seconds with 0 seconds delay, whilst incubating the reaction mixture at 37°C. For each run the spectrometer was first blanked with 1 mL of buffer and 5 µL of the corresponding cell lysate. For Substrate alone, the spectrometer was blanked with 1 mL of buffer. | ||
| + | <br><li><b>Reaction mixture:</b></li> | ||
| + | When the cuvettes were in place in the spectrometer, 4 µL of para-Nitrophenyl butyrate solution was added to each. The reaction mixture was mixed by pipetting up and down and the spectrometer programme was started immediately afterwards.</li></ol> | ||
| + | |||
| + | <h2>3HB assay</h2> | ||
| + | <p>We used a 3-hydroxybutyrate colorimetric assay kit. You can download the description of the product and the protocol [https://www.caymanchem.com/pdfs/700190.pdf here]. The chemical reaction behind the assay is illustrated on the image below:</p> | ||
| + | |||
| + | https://static.igem.org/mediawiki/parts/e/e3/3HB_kit_assay.png | ||
| + | |||
| + | |||
| + | <h4>Sample Preparation: 3HB from P3HB</h4> | ||
| + | <ul> | ||
| + | |||
| + | <p>We treated around 0.1-0.5g of P3HB purified from various experiments (and from Sigma) with 10uL of [http://parts.igem.org/Part:BBa_K1149010 phaZ1] enzyme in 800uL tris buffer. The next day, we centrifuged the samples at 13 000rpm for 10 minutes and used the supernatant in different dilutions with the Assay Buffer (at 90%, 50% and, 10%).</p> | ||
| + | <li><b>Sample 1:</b> Bioplastic made by [http://parts.igem.org/Part:BBa_K1149051 hybrid phaCAB] grown on 3% glucose LB</li> | ||
| + | <li><b>Sample 2:</b> "the grey stuff that is directly from waste". </li> | ||
| + | <li><b>Sample 3:</b> P(3HB) from Sigma ([http://www.sigmaaldrich.com/catalog/product/aldrich/363502?lang=en®ion=GB CAS no. 29435-48-1]) treated with phaZ1</li> | ||
| + | <li><b>Sample 4:</b> P(3HB) from Sigma ([http://www.sigmaaldrich.com/catalog/product/aldrich/363502?lang=en®ion=GB CAS no. 29435-48-1]) untreated, to determine signal from P(3HB)</li> | ||
| + | |||
| + | |||
| + | We did all samples in a 96 well plate) in triplicate. Measurements were taken after 40 minutes incubation with Developer Solution at 20°C. plate setup: | ||
| + | |||
| + | https://static.igem.org/mediawiki/parts/4/40/96_well_plate_3HB.jpg | ||
| + | </ul><br> | ||
| + | |||
| + | <h2>Cloning</h2> | ||
| + | <b>PCR</b> | ||
| + | <ul> | ||
| + | <li>We used Pfu Ultra polymerase for all reactions where we use the DNA for anything afterwards. Download the exact manual from [https://www.chem.agilent.com/Library/usermanuals/Public/600670.pdf here].</li> | ||
| + | |||
| + | <li>We used MyTaq polymerase for colony PCR and checking plasmids. You can download the manual [http://www.bioline.com/documents/product_inserts/MyTaq%E2%84%A2%20DNA%20Polymerase.pdf#zoom=130 here].</li> | ||
| + | |||
| + | <li>Our super Thermal Cycler was provided by our generous sponsor, Eppendorf. </li> | ||
| + | </ul><br> | ||
| + | <b>Running gels</b> | ||
| + | |||
| + | We used SybrSafe to stain the DNA and imaged in a Transilluminator. We cut out bands under a bluebox. | ||
| + | <br><br> | ||
| + | <b>Minipreps</b> | ||
| + | <ul> | ||
| + | We did minipreps from 4mL overnight cultures in LB, supplemented with antibiotics. | ||
| + | <li>Chlo: 50 mg/mL</li> | ||
| + | <li>Amp:100 mg/mL</li> | ||
| + | <li>Kan: 100 mg/mL</li> | ||
| + | <li>We used [http://www.qiagen.com/Products/Catalog/Sample-Technologies/DNA-Sample-Technologies/Plasmid-DNA/QIAprep-Spin-Miniprep-Kit Quiagen Miniprep kit].</li> | ||
| + | |||
| + | We got an excellent tabletop centrifuge courtesy of Eppendorf, which has served us very well. | ||
| + | </ul><br> | ||
| + | <b>Digests and Ligations</b> | ||
| + | <p> For restriction digests, we used [https://www.neb.com/ NEB] enzymes and used the companys protocols accordingly. For ligations, we used T4 ligase. We treated backbones with alkaline phosphatase before ligations and we have used Dpn1 treatment sometimes to degrade the original plasmid. </p> | ||
| + | <br> | ||
| + | <b>Sequencing</b> | ||
| + | <ul> | ||
| + | <li>We used VF2, VR, G1004 and G1005 primers in order to verify the sequence of most of the biobrick parts. We also designed internal sequencing primers to verify the reads in long parts, such as phaCAB. </li> | ||
| + | |||
| + | <li> [http://parts.igem.org/Part:BBa_G00100 VF2 (BBa_G00100)] tgccacctgacgtctaagaa Tm=62</li> | ||
| + | <li> [http://parts.igem.org/Part:BBa_G00100 VR (BBa_G00101)] attaccgcctttgagtgagc Tm=60</li> | ||
| + | <li>[http://parts.igem.org/Part:BBa_G1004 BBa_G1004] (=prefix) gtttcttcgaattcgcggccgcttctag Tm=63</li> | ||
| + | <li>[http://parts.igem.org/Part:BBa_G1004 BBa_G1005] (=revcompl sufix) gtttcttcctgcagcggccgctactagta Tm=64</li> | ||
| + | </ul><br> | ||
| + | <b>Transformation</b> | ||
| + | <p> We transformed our plasmids finally into MG1655. </p> | ||
| + | <p>For ligations we used mainly [https://www.neb.com/products/c3019-neb-10-beta-competent-e-coli-high-efficiency NEB10] and some TOP10 and NEB5 cells for high efficiency. </p> | ||
| + | |||
| + | |||
| + | |||
| + | <h2>Waste Growth Assay</h2> | ||
| + | <p> | ||
| + | Overnight (O/N) cultures of MG1655 transformed with (BBa_K639003) were diluted to OD 0.05 in either fresh LB or waste conditioned media and plated into 96 well plates (200 µl/well). OD600 was read at the indicated time points and media only (LB) was taken away as background signal. | ||
| + | |||
| + | In addition to this a qualitative assay as performed. 9 mL mCherry bacteria were transferred into 300 mL of autoclaved SRF in LB, which were placed in a shaking incubator at 37°C. After 2 days 200 µL of this solution was plated out on chloramphenicol plates, this was then repeated a week later to gauge whether or not the bacteria were still alive. | ||
| + | |||
| + | To show that the MG1655 could grow on waste solely, they were grown in PBS (phosphate buffered saline) and autoclaved waste. In 100 mL PBS + waste, 1 mL MG1655 mCherry E.coli were added. They were then grown in a shaking incubator at 37°C and samples were plated after 2 days and 6 days. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h2>Growth assays</h2> | ||
| + | |||
| + | <h3>3HB and Acetoacetate LB and M9 growth for P3HB synthesis</h3> | ||
| + | <p> | ||
| + | 3HB solutions were prepared in advance. An initial stock of 100mM 3HB was made from 0.315g 3HB (FW:126) dissolved in 25 mL ddH2O. This was then diluted down to 10mM, 1mM, 100µM, 1µM working solutions in LB. Furthermore, 3% glucose in LB was prepared with solutions made from 3g glucose in 100 mL LB. | ||
| + | |||
| + | O/N cultures were prepared, these were diluted to OD=0.05 by measuring O/N OD0.05. With this complete the required dilutions of cells were made into 1.5 mL reaction tubes to give a total volume of 800 µL, this was then divided up into 4 wells for quadruplicate technical repeats. Cells used were phaABC, which is involved in P3HB synthesis from glucose, phaBC which synthesises P3HB from acetoacetate and EV empty vector which was used as a growth control. | ||
| + | <br> | ||
| + | [[File:3HB_table.PNG]] | ||
| + | <br> | ||
| + | With the plate loaded, it was then placed in a shaking incubator at 700 rpm, 37°C for 30 minutes. After this, we took readings using a robotic plate reader for t=0 at OD600, then repeated this over a further 7 hours. | ||
| + | |||
| + | A second plate was prepared to measure acetoacetate as a sole carbon source. Lithium acetoacetate (FW: 108.02) was prepared by placing 10 mg in 9.2 mL water to give a stock concentration of acetoacetate of 10 mM. Dilutions were made to 100 µM and 1µM. Ethylene glycol growth assays were prepared by diluting 1.11 g/L stock solution by 1000 and 500 to give final concentration of 100 mM and 200 mM respectively. Emulsions were prepared as described below. These were further diluted from stock solutions of 1.25 g/L to working solutions of 31 mg/L and 31 µg/L. | ||
| + | <br>[[File:Acetoacetate_table.PNG]] <br> | ||
| + | |||
| + | |||
| + | </p> | ||
| + | |||
| + | |||
| + | <!-- | ||
| + | <h2>[http://biolabs.tmcc.edu/Micro%20Web/SerialDilutions.pdf Serial Dilution]</h2> | ||
| + | --> | ||
| + | |||
| + | <h2>Preparation of Poly DEGA plates.</h2> | ||
| + | <p> | ||
| + | For 320 mL final media, place 1.5 mL 0.5% poly DEGA in 18.5 mL ddH20. As poly DEGA is very viscous, the 20 mL solution was sonicated for 45 minutes. This complete, it was added to LB agar kept at 70°C along with 25 ug/mL chloramphenicol. The solution was then separated into 3 separate duran bottles of 100 mL volume. 0.21g xylose was added to a bottle, 0.21g arabinose to another and the other did not have an inducer added. The media was then plated out. | ||
| + | </p> | ||
| + | |||
| + | <h2>Western blot</h2> | ||
| + | <p><b> | ||
| + | Sample preparation | ||
| + | </b></p> | ||
| + | <ol> | ||
| + | |||
| + | <li>Prepare concentrated induction media. (Dissolve 0.134g Arabinose in 8ml pre-autoclaved LB, 1.52g xylose in 4ml pre-autoclaved LB). Filter sterilise induction media. </li> | ||
| + | <li> Make 5 mL overnight culture from the transformed cells. | ||
| + | </li> | ||
| + | <li> Next morning, dilute 0.5 ml overnight culture in 9.5 ml of LB media. The subsequent inducer concentrations are 6mM arabinose and 2% xylose. | ||
| + | </li> | ||
| + | <li> Then grow the diluted culture at 37°C, and measure OD600 every hour until the OD reaches 0.5-0.6. | ||
| + | </li> | ||
| + | <li> Then induce protein expression with the appropriate induction media, and grow the induced cells at 37°C for 5 hours. | ||
| + | </li> | ||
| + | <li> Spin 2ml of induced cells, collect 60ul of the supernatant with a 1.5ml tube, and add 20 ul western loading buffer to the supernatant. | ||
| + | </li> | ||
| + | <li> Resuspend the pellet with 100 ul of lysis buffer. Wait for the cells to lyse (15-20 minutes) at room temperature. Vortex is allowed. | ||
| + | </li> | ||
| + | <li> Add 33 ul of western loading buffer to the lysate. | ||
| + | </li> | ||
| + | <li> Boil the sample at 95°C for 5 minutes and freeze at -20°C. | ||
| + | </li></ol> | ||
| + | |||
| + | <b> Gel electrophoresis </b> | ||
| + | <ul> | ||
| + | <li>Place 15ul of the sample in each well. Run the gel at 60V for 1 hour and 20 minutes.</li> | ||
| + | </ul> | ||
| + | |||
| + | <b> Transfer </b> | ||
| + | <ul> | ||
| + | <li>Use [http://www.lifetechnologies.com/uk/en/home/life-science/protein-expression-and-analysis/western-blotting/western-blot-transfer/iblot-dry-blotting-system.html iBlot® Dry Blotting System (Invitrogen)] to perform dry blotting. | ||
| + | </li> | ||
| + | </ul> | ||
| + | |||
| + | <b> Antibody binding</b> | ||
| + | |||
| + | All-in-one his antibody (from mouse) is used for proteins with his-tags. Primary and secondary Flag-antibodies are used for Flag-tagged proteins. | ||
| + | Mix 7 ml of 5% w/v non-fat dry milk with diluted antibodies, and place the membrane in the mixture. | ||
| + | Incubate for 1 hour at room temperature. | ||
| + | Wash 210 min with 15 ml 0.05% PHB-Tween. | ||
| + | Wash 25 min with 15 ml PBS. | ||
| + | </p> | ||
| + | <p><b> | ||
| + | Visualisation | ||
| + | </b></p> | ||
| + | <p> | ||
| + | Visualise the protein bands using [http://www.lifetechnologies.com/1/1/28062-novex-ap-mouse-chemiluminescent-detection-kit.html Chemiluminescence detection kit (Invitrogen)]. Protein-specific signal is then captured by a chemiluminescent-compatible X-ray film. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h2>Clearing zone assay</h2> | ||
| + | <ol> | ||
| + | <li>Prepare 10 ml cell culture and induce phaZ1 expression using 2% xylose media. </li> | ||
| + | <li>Centrifuge the cell cultures at 4500 RCF (equivalent to 6221g) for 20 minutes at 4°C.</li> | ||
| + | <li>Add 2 ml of 50mM Tris buffer (pH 7.4) to the pellet and mix well by vortexing the tube. </li> | ||
| + | <li>Transfer the buffered pellet into 7ml tubes and keep the samples on ice.</li> | ||
| + | <li>Perform probe sonication with 2-minute cycles of 2 seconds on, 2 seconds off, at 60% amplitude. Keep the sample on ice while sonicating.</li> | ||
| + | <li>Centrifuge the samples again at 4000 g for 10 minutes and extract the supernatant, in order to separate cell debris and proteins.</li> | ||
| + | <li>Mix 0.5g of P3HB powder with 300 ml LB agar, make P3HB plates.</li> | ||
| + | <li>Use sterilised pipette tip to cut two wells, each has a 0.5 mm diameter. </li> | ||
| + | <li>Pipette 200ul of PhaZ1 cell lysate sample in one of the wells, and same amount of empty vector cell lysate sample in the other well.</li> | ||
| + | <li>Observe the plate every 24 hours.</li> | ||
| + | </ol> | ||
| + | |||
| + | |||
| + | <h2>Protein purification by metal affinity chromatography</h2> | ||
| + | <ol> | ||
| + | <li><b>Preparation:</b></li> | ||
| + | 10 mL cultures were incubated overnight. These were diluted in 500 mL the next morning and incubated until the OD reached 0.5-0.6. The cultures were then induced with 6mM arabinose or 2% xylose (final concentration)and left overnight. The cells were then pelleted by centrifugation, 4,000 rpm for 10 minutes and kept in a -80ºC freezer. | ||
| + | |||
| + | <br><li><b>Cell lysis</b></li> | ||
| + | The frozen pellets were allowed to thaw and resuspended in 10 mL of Tris-HCl binding buffer. The cells were lysed by probe-sonication at 60% amplitude with 2 minute cycles of 2 seconds on, 2 seconds off. The samples were kept on ice throughout the sonication process. The cell debris was pelleted by spinning and the supernatant removed for purification. | ||
| + | |||
| + | <br><li><b>Purification with a Ni-NTA column:</b></li> | ||
| + | The column was washed with Buffer A (10 mM imidazole, 40 mM Tris pH 8, and 400 mM NaCl) to equilibrate the resin. The samples are added and the column is washed with Buffer A, and then eluted with increasing concentrations of Buffer B (500 mM imidazole, 40 mM Tris pH 8 and 400 mM NaCl). | ||
| + | |||
| + | <br><li><b>SDS PAGE:</b></li> | ||
| + | Samples were prepared for SDS PAGE by adding 7 µL of SDS running buffer to 20 µL of the fractions of interest. 8-20 µL of each sample were run on a polyacrylamide gel against a molecular weight marker. | ||
| + | |||
| + | <br><li><b>Concentration:</b></li> | ||
| + | [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149010 PHB depolymerase (phaz 1)] and [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149002 PUR esterase (EstC2)] were concentrated with 10 mM potassium phosphate buffer. | ||
| + | |||
| + | <h2>Proteinase K kinetic activity assay</h2> | ||
| - | + | <ol> | |
| + | <li><b>Tris-HCl buffer ingredients:</b></li> | ||
| + | 1 litre of millicule water | ||
| + | 10.5 g of NaCl (180 mM) | ||
| + | 0.65 g of CaCl2 (5 mM) | ||
| + | 1.7g of imidazole (25 mM) | ||
| + | 6 g of Tris base (50 mM) | ||
| + | Add HCl to bring the pH to 7.4 | ||
| - | <br> < | + | <br><li><b>Programme the spectrometer:</b></li> |
| - | <b> | + | Cuvette: 10 mm |
| - | < | + | Wavelength: 405 nm |
| + | Temperature: 37°C | ||
| + | Measuring time: 10:00 min:sec | ||
| + | Delay: 00:00 min:sec | ||
| + | Interval: 00:30 min:sec | ||
| - | <b> | + | <br><li><b>Reagent preparation:</b></li> |
| - | < | + | Lyophilized powder proteinase K from Tritirachium album from Sigma-Aldrich was used, made into a 10 mg/ml stock solution with the buffer. The spectrometer was blanked with 1mL of buffer and 3 µL of proteinase K stock solution; this is equivalent to one unit of enzyme. |
| + | |||
| + | We used N-Succinyl Ala-Ala-Pro-Leu p-nitroanilide (AAPLp-NA) from Sigma-Aldrich as a substrate analog. Make a stock solution with buffer (10 mg/ml). The reagents were incubated at 37°C just before use. | ||
| + | The absorbance of each reaction mixture was measured at 405 nm, for 10 minutes at 30 second intervals. The reaction mixtures contained 3 µL of proteinase K stock solution and AAPLp-NA in Tris-HCl buffer to make final concentrations of 10 µM, 25 µM, 50 µM, 75 µM, 100 µM, 250 µM and 500 µM. | ||
| - | + | {{:Team:Imperial_College/Templates:footer}} | |
Latest revision as of 02:43, 5 October 2013
Materials
Waste Conditioned Media (WCM)
We added 1g [http://en.wikipedia.org/wiki/Refuse-derived_fuel SRF] (Solid Recovered Fuel) /50 mL LB and then autoclaved the mixture. Once autoclaved, large waste chunks were removed through filter sterilisation (0.2 µm filters) before being used in growth assays as waste conditioned media (WCM). SRF refers to the refuse from recycling facilities that has no value currently and is incinerated to produce power at a cost to the recycling facility, it is composed of 30% plastics while the rest is cellulosic waste
3% glucose Nile Red plates
When making up LB agar media, add 3% glucose then autoclave. When autoclaving it is essential that you keep temperature <125°C. Failure to do so will result in caramelisation (160°C) of the media, which will show as a darkening of the media. With this complete, 60 µL Nile Red was added in addition to 150 µL chloramphenicol in a 300 mL final volume. Plates were then poured and left to dry. Another successful mechanism to thwart caramelisation is to autoclave the LB-agar then add the 3% glucose after. Follow this with a 30 second microwave step and then an 80°C water bath for a couple hours to thoroughly sterilise.
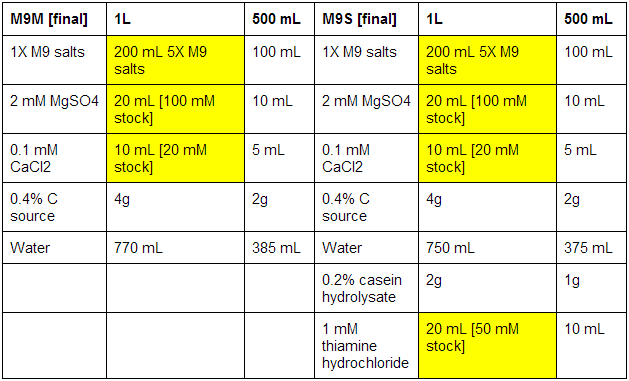
M9 minimal and supplemented media
First made stocks of MgSO4 (246.48 g/mol), CaCl2 (147.02 g/mol), and thiamine hydrochloride (337.3 g/mol):
MgSO4: 100 mM - 2.46g in 100 mL ddH2O
CaCl2: 20 mM - 0.29g in 100 mL ddH2O
Thiamine hydrochloride: 50 mM - 0.84g in 50 mL ddH2O
The bottles used were pre-autoclaved. When all media was prepared, it was sterile filtrated, as autoclaving results in the formation of magnesium precipitate. See below for correct volumes and mass to add where [final] represents desired values.
Xylose and Arabinose
For induction assays, 2% xylose was prepared, as was 6 µM arabinose.
Protocols and Assays

P3HB production and extraction
Extracting PHB
- Centrifuge settings: 4000RPM, 10mins.
- Scale as appropriate.
- After each centrifuge step the supernatant should be poured off.
- This should provide PHB with 99% purity and a high molecular weight.
- Centrifuge 50ml culture and resuspend in PBS.
- Resuspend pellet in 5ml Triton X-100(1% v/v in PBS) for 30mins at room temp.
- Centrifuge, resuspend in 5ml PBS.
- Centrifuge, add 5ml sodium hyperchlorite solution and incubate at 30˚C for 1 hour.
- Centrifuge, wash with 5ml distilled water and 70% EtOH several times.
- Allow powder to dry.
Shahryar Shakeri, Comparison of intracellular polyhydroxybutyrate granules formation between different bacterial cell subpopulations by flow cytometry, Jundishapur Journal of Microbiology 2011
[http://jjmicrobiol.com/?page=article&article_id=2342]
LB-Agar Plates containing Nile Red stain
Nile Red final concentration in media should be 0.5µg/ml
- After autoclaving 300ml of LB-Agar, add 600µl Nile Red solution(0.25mg/ml DMSO)
- Add 150µl chloramphenicol
- Pour plates in a clean fume cupboard
Nile Red Culture staining
Nile Red final concentration should be 20µg/ml media
Add 320µl to 4 ml overnight culture and incubate for 30mins at room temp.
Fluorescent Microscopy
Agar pads are first made
- Small pads are made using 1% agarose in M9 minimal media between microscope slides.
- 2ul of the sample is added to each pad and imaged with the fluorescent microscope.
Relationship between OD and dry biomass for phaCAB operon containing cells
- Grow 3 large cultures in LB and 3% glucose - inoculate with 1ml OD 0.1 and grow for 24h.
- Calculate OD of original culture and dilute until there is a range of 0.2 - 1.0 OD(try to have large volumes for low OD).
- Weigh centrifuge containers.
- Transfer solutions to these containers, recording what volume of each OD is transferred. .
- Centrifuge these solutions down and dry the pellet out until constant weight in a 50°C oven.
- Dry cell mass = (container mass + dry cell mass)/container mass
- Plot OD against dry cell mass.
Cell lysate activity assay
- Preparation of the cell lysate:
- Substrate preparation:
- Incubation of the reagents:
- Spectrometer programme:
- Reaction mixture: When the cuvettes were in place in the spectrometer, 4 µL of para-Nitrophenyl butyrate solution was added to each. The reaction mixture was mixed by pipetting up and down and the spectrometer programme was started immediately afterwards.
5 mL cultures of EstC2 [http://parts.igem.org/Part:BBa_K1149002 BBa_K1149002], PueB [http://parts.igem.org/Part:BBa_K1149004 BBa_K1149004] and PulA [http://parts.igem.org/Part:BBa_K1149006 BBa_K1149006] were incubated overnight. The next morning, these were diluted in 200 mL and incubated until the OD reached 0.5-0.6.
The cultures were then induced with 6mM arabinose (final concentration)and left overnight. The following morning the cells were spun down, resuspended in 100 mM potassium phosphate buffer of pH 7.4 and probe-sonicated with 2 minute cycles of 2 seconds on, 2 seconds off, at 60% amplitude. The samples were kept on ice throughout the sonication process. The cell debris was pelleted by spinning and the supernatant was removed for utilising in the enzyme activity assays.The substrate was prepared by the addition of 8.8 µL of para-Nitrophenyl butyrate to 1 mL of Acetonitril.
In 10mm cuvettes, 5 µL of the EstC2, PueB, PulA, and Empty vector were added to 1 mL of 100 mM Potassium Phosphate buffer of pH 7.4 and incubated for 2-3 minutes. For the Substrate alone assay, simply 1 mL of buffer was incubated.
The Eppendorf BioSpectrometer was set up to run for 600 seconds, recording the absorbance at 405 nm every 30 seconds with 0 seconds delay, whilst incubating the reaction mixture at 37°C. For each run the spectrometer was first blanked with 1 mL of buffer and 5 µL of the corresponding cell lysate. For Substrate alone, the spectrometer was blanked with 1 mL of buffer.
3HB assay
We used a 3-hydroxybutyrate colorimetric assay kit. You can download the description of the product and the protocol here. The chemical reaction behind the assay is illustrated on the image below:

Sample Preparation: 3HB from P3HB
- Sample 1: Bioplastic made by [http://parts.igem.org/Part:BBa_K1149051 hybrid phaCAB] grown on 3% glucose LB
- Sample 2: "the grey stuff that is directly from waste".
- Sample 3: P(3HB) from Sigma ([http://www.sigmaaldrich.com/catalog/product/aldrich/363502?lang=en®ion=GB CAS no. 29435-48-1]) treated with phaZ1
- Sample 4: P(3HB) from Sigma ([http://www.sigmaaldrich.com/catalog/product/aldrich/363502?lang=en®ion=GB CAS no. 29435-48-1]) untreated, to determine signal from P(3HB)
We treated around 0.1-0.5g of P3HB purified from various experiments (and from Sigma) with 10uL of [http://parts.igem.org/Part:BBa_K1149010 phaZ1] enzyme in 800uL tris buffer. The next day, we centrifuged the samples at 13 000rpm for 10 minutes and used the supernatant in different dilutions with the Assay Buffer (at 90%, 50% and, 10%).
We did all samples in a 96 well plate) in triplicate. Measurements were taken after 40 minutes incubation with Developer Solution at 20°C. plate setup:

Cloning
PCR
- We used Pfu Ultra polymerase for all reactions where we use the DNA for anything afterwards. Download the exact manual from here.
- We used MyTaq polymerase for colony PCR and checking plasmids. You can download the manual [http://www.bioline.com/documents/product_inserts/MyTaq%E2%84%A2%20DNA%20Polymerase.pdf#zoom=130 here].
- Our super Thermal Cycler was provided by our generous sponsor, Eppendorf.
Running gels
We used SybrSafe to stain the DNA and imaged in a Transilluminator. We cut out bands under a bluebox.
Minipreps
-
We did minipreps from 4mL overnight cultures in LB, supplemented with antibiotics.
- Chlo: 50 mg/mL
- Amp:100 mg/mL
- Kan: 100 mg/mL
- We used [http://www.qiagen.com/Products/Catalog/Sample-Technologies/DNA-Sample-Technologies/Plasmid-DNA/QIAprep-Spin-Miniprep-Kit Quiagen Miniprep kit].
We got an excellent tabletop centrifuge courtesy of Eppendorf, which has served us very well.
Digests and Ligations
For restriction digests, we used NEB enzymes and used the companys protocols accordingly. For ligations, we used T4 ligase. We treated backbones with alkaline phosphatase before ligations and we have used Dpn1 treatment sometimes to degrade the original plasmid.
Sequencing
- We used VF2, VR, G1004 and G1005 primers in order to verify the sequence of most of the biobrick parts. We also designed internal sequencing primers to verify the reads in long parts, such as phaCAB.
- [http://parts.igem.org/Part:BBa_G00100 VF2 (BBa_G00100)] tgccacctgacgtctaagaa Tm=62
- [http://parts.igem.org/Part:BBa_G00100 VR (BBa_G00101)] attaccgcctttgagtgagc Tm=60
- [http://parts.igem.org/Part:BBa_G1004 BBa_G1004] (=prefix) gtttcttcgaattcgcggccgcttctag Tm=63
- [http://parts.igem.org/Part:BBa_G1004 BBa_G1005] (=revcompl sufix) gtttcttcctgcagcggccgctactagta Tm=64
Transformation
We transformed our plasmids finally into MG1655.
For ligations we used mainly NEB10 and some TOP10 and NEB5 cells for high efficiency.
Waste Growth Assay
Overnight (O/N) cultures of MG1655 transformed with (BBa_K639003) were diluted to OD 0.05 in either fresh LB or waste conditioned media and plated into 96 well plates (200 µl/well). OD600 was read at the indicated time points and media only (LB) was taken away as background signal. In addition to this a qualitative assay as performed. 9 mL mCherry bacteria were transferred into 300 mL of autoclaved SRF in LB, which were placed in a shaking incubator at 37°C. After 2 days 200 µL of this solution was plated out on chloramphenicol plates, this was then repeated a week later to gauge whether or not the bacteria were still alive. To show that the MG1655 could grow on waste solely, they were grown in PBS (phosphate buffered saline) and autoclaved waste. In 100 mL PBS + waste, 1 mL MG1655 mCherry E.coli were added. They were then grown in a shaking incubator at 37°C and samples were plated after 2 days and 6 days.
Growth assays
3HB and Acetoacetate LB and M9 growth for P3HB synthesis
3HB solutions were prepared in advance. An initial stock of 100mM 3HB was made from 0.315g 3HB (FW:126) dissolved in 25 mL ddH2O. This was then diluted down to 10mM, 1mM, 100µM, 1µM working solutions in LB. Furthermore, 3% glucose in LB was prepared with solutions made from 3g glucose in 100 mL LB.
O/N cultures were prepared, these were diluted to OD=0.05 by measuring O/N OD0.05. With this complete the required dilutions of cells were made into 1.5 mL reaction tubes to give a total volume of 800 µL, this was then divided up into 4 wells for quadruplicate technical repeats. Cells used were phaABC, which is involved in P3HB synthesis from glucose, phaBC which synthesises P3HB from acetoacetate and EV empty vector which was used as a growth control.
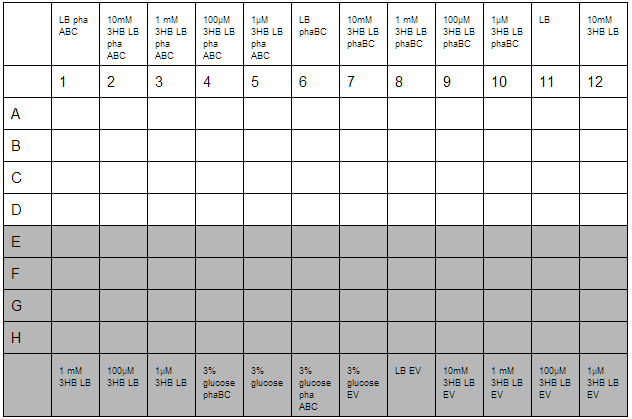
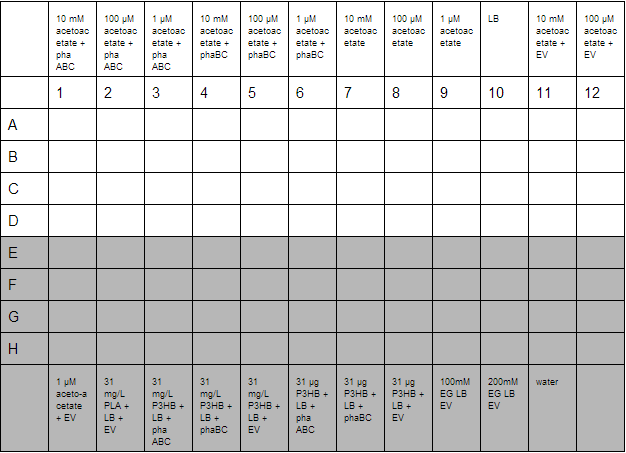
With the plate loaded, it was then placed in a shaking incubator at 700 rpm, 37°C for 30 minutes. After this, we took readings using a robotic plate reader for t=0 at OD600, then repeated this over a further 7 hours.
A second plate was prepared to measure acetoacetate as a sole carbon source. Lithium acetoacetate (FW: 108.02) was prepared by placing 10 mg in 9.2 mL water to give a stock concentration of acetoacetate of 10 mM. Dilutions were made to 100 µM and 1µM. Ethylene glycol growth assays were prepared by diluting 1.11 g/L stock solution by 1000 and 500 to give final concentration of 100 mM and 200 mM respectively. Emulsions were prepared as described below. These were further diluted from stock solutions of 1.25 g/L to working solutions of 31 mg/L and 31 µg/L.
Preparation of Poly DEGA plates.
For 320 mL final media, place 1.5 mL 0.5% poly DEGA in 18.5 mL ddH20. As poly DEGA is very viscous, the 20 mL solution was sonicated for 45 minutes. This complete, it was added to LB agar kept at 70°C along with 25 ug/mL chloramphenicol. The solution was then separated into 3 separate duran bottles of 100 mL volume. 0.21g xylose was added to a bottle, 0.21g arabinose to another and the other did not have an inducer added. The media was then plated out.
Western blot
Sample preparation
- Prepare concentrated induction media. (Dissolve 0.134g Arabinose in 8ml pre-autoclaved LB, 1.52g xylose in 4ml pre-autoclaved LB). Filter sterilise induction media.
- Make 5 mL overnight culture from the transformed cells.
- Next morning, dilute 0.5 ml overnight culture in 9.5 ml of LB media. The subsequent inducer concentrations are 6mM arabinose and 2% xylose.
- Then grow the diluted culture at 37°C, and measure OD600 every hour until the OD reaches 0.5-0.6.
- Then induce protein expression with the appropriate induction media, and grow the induced cells at 37°C for 5 hours.
- Spin 2ml of induced cells, collect 60ul of the supernatant with a 1.5ml tube, and add 20 ul western loading buffer to the supernatant.
- Resuspend the pellet with 100 ul of lysis buffer. Wait for the cells to lyse (15-20 minutes) at room temperature. Vortex is allowed.
- Add 33 ul of western loading buffer to the lysate.
- Boil the sample at 95°C for 5 minutes and freeze at -20°C.
Gel electrophoresis
- Place 15ul of the sample in each well. Run the gel at 60V for 1 hour and 20 minutes.
Transfer
- Use [http://www.lifetechnologies.com/uk/en/home/life-science/protein-expression-and-analysis/western-blotting/western-blot-transfer/iblot-dry-blotting-system.html iBlot® Dry Blotting System (Invitrogen)] to perform dry blotting.
Antibody binding
All-in-one his antibody (from mouse) is used for proteins with his-tags. Primary and secondary Flag-antibodies are used for Flag-tagged proteins. Mix 7 ml of 5% w/v non-fat dry milk with diluted antibodies, and place the membrane in the mixture. Incubate for 1 hour at room temperature. Wash 210 min with 15 ml 0.05% PHB-Tween. Wash 25 min with 15 ml PBS. </p>
Visualisation
Visualise the protein bands using [http://www.lifetechnologies.com/1/1/28062-novex-ap-mouse-chemiluminescent-detection-kit.html Chemiluminescence detection kit (Invitrogen)]. Protein-specific signal is then captured by a chemiluminescent-compatible X-ray film.
Clearing zone assay
- Prepare 10 ml cell culture and induce phaZ1 expression using 2% xylose media.
- Centrifuge the cell cultures at 4500 RCF (equivalent to 6221g) for 20 minutes at 4°C.
- Add 2 ml of 50mM Tris buffer (pH 7.4) to the pellet and mix well by vortexing the tube.
- Transfer the buffered pellet into 7ml tubes and keep the samples on ice.
- Perform probe sonication with 2-minute cycles of 2 seconds on, 2 seconds off, at 60% amplitude. Keep the sample on ice while sonicating.
- Centrifuge the samples again at 4000 g for 10 minutes and extract the supernatant, in order to separate cell debris and proteins.
- Mix 0.5g of P3HB powder with 300 ml LB agar, make P3HB plates.
- Use sterilised pipette tip to cut two wells, each has a 0.5 mm diameter.
- Pipette 200ul of PhaZ1 cell lysate sample in one of the wells, and same amount of empty vector cell lysate sample in the other well.
- Observe the plate every 24 hours.
Protein purification by metal affinity chromatography
- Preparation:
- Cell lysis
- Purification with a Ni-NTA column:
- SDS PAGE:
- Concentration:
- Tris-HCl buffer ingredients:
- Programme the spectrometer:
- Reagent preparation:
10 mL cultures were incubated overnight. These were diluted in 500 mL the next morning and incubated until the OD reached 0.5-0.6. The cultures were then induced with 6mM arabinose or 2% xylose (final concentration)and left overnight. The cells were then pelleted by centrifugation, 4,000 rpm for 10 minutes and kept in a -80ºC freezer.
The frozen pellets were allowed to thaw and resuspended in 10 mL of Tris-HCl binding buffer. The cells were lysed by probe-sonication at 60% amplitude with 2 minute cycles of 2 seconds on, 2 seconds off. The samples were kept on ice throughout the sonication process. The cell debris was pelleted by spinning and the supernatant removed for purification.
The column was washed with Buffer A (10 mM imidazole, 40 mM Tris pH 8, and 400 mM NaCl) to equilibrate the resin. The samples are added and the column is washed with Buffer A, and then eluted with increasing concentrations of Buffer B (500 mM imidazole, 40 mM Tris pH 8 and 400 mM NaCl).
Samples were prepared for SDS PAGE by adding 7 µL of SDS running buffer to 20 µL of the fractions of interest. 8-20 µL of each sample were run on a polyacrylamide gel against a molecular weight marker.
[http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149010 PHB depolymerase (phaz 1)] and [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149002 PUR esterase (EstC2)] were concentrated with 10 mM potassium phosphate buffer.
Proteinase K kinetic activity assay
1 litre of millicule water 10.5 g of NaCl (180 mM) 0.65 g of CaCl2 (5 mM) 1.7g of imidazole (25 mM) 6 g of Tris base (50 mM) Add HCl to bring the pH to 7.4
Cuvette: 10 mm Wavelength: 405 nm Temperature: 37°C Measuring time: 10:00 min:sec Delay: 00:00 min:sec Interval: 00:30 min:sec
Lyophilized powder proteinase K from Tritirachium album from Sigma-Aldrich was used, made into a 10 mg/ml stock solution with the buffer. The spectrometer was blanked with 1mL of buffer and 3 µL of proteinase K stock solution; this is equivalent to one unit of enzyme.
We used N-Succinyl Ala-Ala-Pro-Leu p-nitroanilide (AAPLp-NA) from Sigma-Aldrich as a substrate analog. Make a stock solution with buffer (10 mg/ml). The reagents were incubated at 37°C just before use.
The absorbance of each reaction mixture was measured at 405 nm, for 10 minutes at 30 second intervals. The reaction mixtures contained 3 µL of proteinase K stock solution and AAPLp-NA in Tris-HCl buffer to make final concentrations of 10 µM, 25 µM, 50 µM, 75 µM, 100 µM, 250 µM and 500 µM.
 "
"



