Team:Goettingen/Team/DAC
From 2013.igem.org
FMCommmichau (Talk | contribs) |
(→Results and Discussion) |
||
| (7 intermediate revisions not shown) | |||
| Line 57: | Line 57: | ||
The truncated DacA protein ([http://parts.igem.org/Part:BBa_K1045003 BBa_K1045003]) was extended with an N-terminal ''Step''-tag allowing the rapid purification using the ''Strep''-tag:Streptactin purification system. Synthesis of our protein is driven by a T7 promoter. This promoter is recognized by the T7 polymerase, which is encoded in the genome of the <i>E. coli</i> strain BL21. Synthesis of the T7 polymerase can be controlled by Isopropyl-β-D-thiogalactopyranoside (IPTG). The generated plasmid was then used to transform the <i>E. coli</i> strain BL21, which is a powerful strain for synthesis of recombinant proteins. In contrast to Gram-positive bacteria, the Gram-negative bacterium ''E. coli'' does not produce c-di-AMP and growth is not affected by the signaling molecule. | The truncated DacA protein ([http://parts.igem.org/Part:BBa_K1045003 BBa_K1045003]) was extended with an N-terminal ''Step''-tag allowing the rapid purification using the ''Strep''-tag:Streptactin purification system. Synthesis of our protein is driven by a T7 promoter. This promoter is recognized by the T7 polymerase, which is encoded in the genome of the <i>E. coli</i> strain BL21. Synthesis of the T7 polymerase can be controlled by Isopropyl-β-D-thiogalactopyranoside (IPTG). The generated plasmid was then used to transform the <i>E. coli</i> strain BL21, which is a powerful strain for synthesis of recombinant proteins. In contrast to Gram-positive bacteria, the Gram-negative bacterium ''E. coli'' does not produce c-di-AMP and growth is not affected by the signaling molecule. | ||
| - | In order to analyze the DAC activity ''in vivo'', the ''E. coli'' clones | + | In order to analyze the DAC activity ''in vivo'', DAC production by the ''E. coli'' clones was induced by adding IPTG. The cells were then lysed to extract c-di-AMP from the cells. |
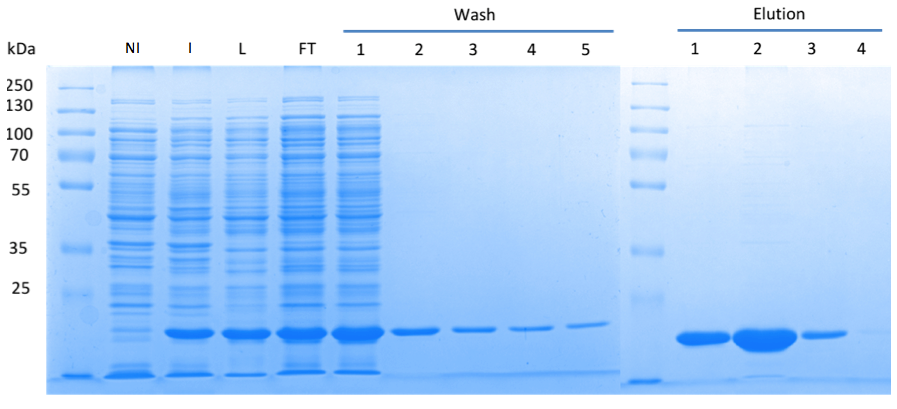
By performing the SDS PAGE, we could nicely show that the desired protein was well-produced (Fig. 1). Furthermore, the presence of c-di-AMP in the supernatant of the lysed bacteria was confirmed using LC-MS/MS. Thus, we can conclude that the truncated DacA protein codes for an DAC domain that is active ''in vivo''. | By performing the SDS PAGE, we could nicely show that the desired protein was well-produced (Fig. 1). Furthermore, the presence of c-di-AMP in the supernatant of the lysed bacteria was confirmed using LC-MS/MS. Thus, we can conclude that the truncated DacA protein codes for an DAC domain that is active ''in vivo''. | ||
| Line 75: | Line 75: | ||
| - | Finally, we are coming to the core of our project, the protein structure of DacA DAC domain! It is very helpful to have the molecular structure of a DAC in hands because it allows to perform ''in silico'' experiments. These ''in silico'' experiments can lead to the discovery of new antibacterial substance classes, that specifically inhibit DAC activity. The final goal is to use potential antibiotics to treat patients that suffer from infection with pathogenic Gram-positive bacteria. | + | Finally, we are coming to the core of our project, the protein structure of the DacA DAC domain! It is very helpful to have the molecular structure of a DAC in hands because it allows to perform ''in silico'' experiments. These ''in silico'' experiments can lead to the discovery of new antibacterial substance classes, that specifically inhibit DAC activity. The final goal is to use potential antibiotics to treat patients that suffer from infection with pathogenic Gram-positive bacteria. |
| - | In order to purify a large amount of this protein, our BioBrick [http://partsregistry.org/Part:BBa_K1045003 BBa_K1045003] | + | In order to purify a large amount of this protein, we used our BioBrick [http://partsregistry.org/Part:BBa_K1045003 BBa_K1045003] to produce the N-terminally ''Strep''-tagged DAC domain in ''E. coli''. Expression was confirmed by SDS PAGE and the protein showed a molecular weight of about 20 kDa (Fig. 3). The ''E. coli'' strain was grown in a large scale (10 liters), the cells were harvested by centrifugation, lysed and the protein was purified by affinity purification (Fig. 3). |
<html><a href="https://static.igem.org/mediawiki/2013/f/fe/Goe-dac-fig-3.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/f/fe/Goe-dac-fig-3.png" width="750" /></a></html> | <html><a href="https://static.igem.org/mediawiki/2013/f/fe/Goe-dac-fig-3.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/f/fe/Goe-dac-fig-3.png" width="750" /></a></html> | ||
| - | Fig. 3. '''SDS | + | Fig. 3. '''SDS PAGE showing high-level production and purification of the DAC domain.''' (A) Lane 1: Thermo Scientific PageRuler Plus Prestained Protein Ladder; NI: Non-induced clone, the cells did not produce the DAC domain and c-di-AMP; I: Induction of ''dac'' expression with IPTG, the clone synthesized the DAC domain and c-di-AMP was detected; L: Lysate; FT: Flow-through; the protein eluted in the first elution fraction. |
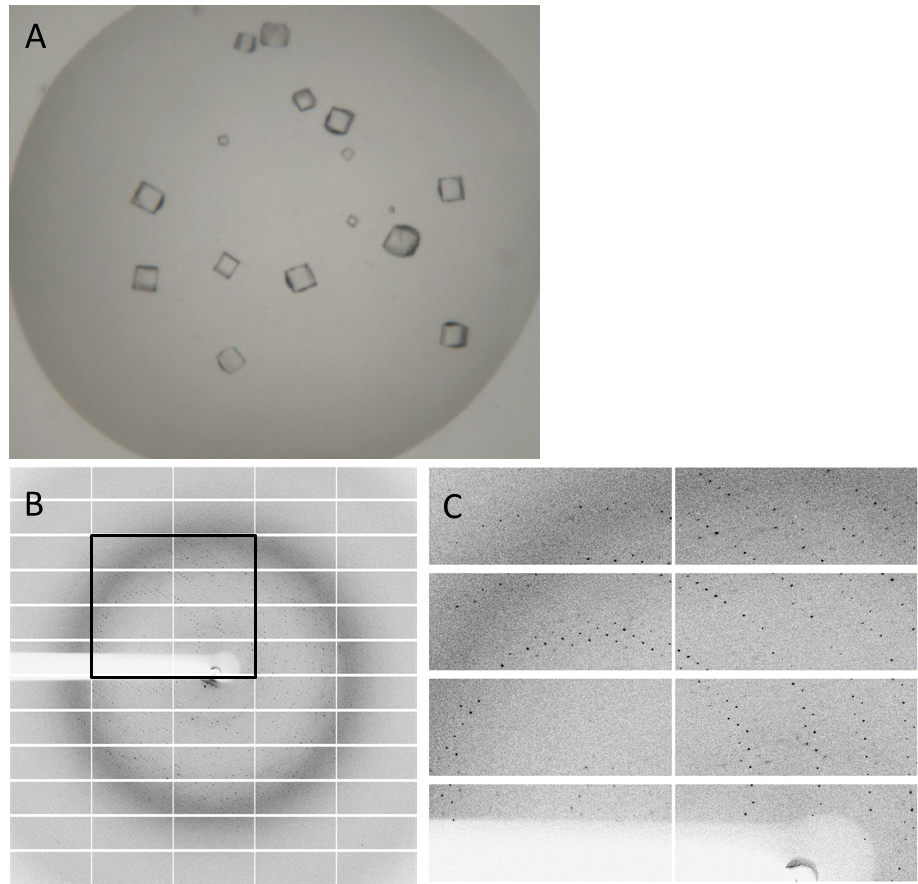
| - | Having dialyzed and concentrated the | + | Having dialyzed and concentrated the purified protein, we obtained a protein solution with a concentration of about 10 mg/ml, which was sufficient to perform a crystallization screening. Luckily, we have obtained very nice crystals in the initial crystallization screen (Fig. 4A). In order to find the perfect supplements for growing our crystals, the whole procedure was repeated and the conditions were refined. The crystals yielded an x-ray diffraction pattern, with a resolution of 2,8 Å (Fig. 4B,C). The dataset was measured at the EMBL Hamburg Beamline P13 at the PETRA III synchrotron on the DESY campus, using a PILATUS2 6M X-ray detector (https://www.dectris.com/). |
Finally, we were also able to obtain the protein structure (Fig. 5)! The structure shows a globular protein with a distinct ATP-binding cleft. The ribbon model demonstrates the secondary structure composed of α-helices and β-sheets. | Finally, we were also able to obtain the protein structure (Fig. 5)! The structure shows a globular protein with a distinct ATP-binding cleft. The ribbon model demonstrates the secondary structure composed of α-helices and β-sheets. | ||
| Line 89: | Line 89: | ||
<html><a href="https://static.igem.org/mediawiki/2013/a/a8/Goe-dac-fig-4.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/a/a8/Goe-dac-fig-4.png" width="750"></a></html> | <html><a href="https://static.igem.org/mediawiki/2013/a/a8/Goe-dac-fig-4.png" target="_blank"><img src="https://static.igem.org/mediawiki/2013/a/a8/Goe-dac-fig-4.png" width="750"></a></html> | ||
| - | Fig. 4. '''Crystals and diffraction pattern.''' Nice crystals were | + | Fig. 4. '''Crystals and diffraction pattern.''' Nice crystals were obtained with a medium concentration of alcohol and other supplements (confidential :-]) '''(A).''' X-ray diffraction image of the DacA DAC domain crystals '''(B)'''; the highlighted box is shown enlarged '''(C)'''. The dataset was measured at the EMBL Hamburg Beamline P13 in the PETRA III synchrotron on the DESY campus. |
https://static.igem.org/mediawiki/2013/3/36/Goe-dac-fig-5.png | https://static.igem.org/mediawiki/2013/3/36/Goe-dac-fig-5.png | ||
| - | Fig. 6.''' Protein structure of DacA.''' (A, B) Ribbon | + | Fig. 6.''' Protein structure of the DacA DAC domain.''' (A, B) Ribbon model of the DAC domain in its ATP-bound state. (C, D) Surface structure of the DAC domain and the ATP-binding pocket. (E) Magnified view into the ATP-binding pocket |
| + | <html><object width="420" height="315"><param name="movie" value="//www.youtube.com/v/9BQOEIVsF-Y?hl=zh_CN&version=3&rel=0"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="//www.youtube.com/v/9BQOEIVsF-Y?hl=zh_CN&version=3&rel=0" type="application/x-shockwave-flash" width="420" height="315" allowscriptaccess="always" allowfullscreen="true"></embed></object></html> | ||
'''References''' | '''References''' | ||
Latest revision as of 09:45, 28 October 2013
 "
"