Team:Peking
From 2013.igem.org
XingjiePan (Talk | contribs) |
XingjiePan (Talk | contribs) |
||
| (16 intermediate revisions not shown) | |||
| Line 152: | Line 152: | ||
#DataPage_Sublist{position:relative; top:0px; left:-60px;} | #DataPage_Sublist{position:relative; top:0px; left:-60px;} | ||
#Safety_Sublist{position:relative; top:0px; left:-180px;} | #Safety_Sublist{position:relative; top:0px; left:-180px;} | ||
| - | #HumanPractice_Sublist{position:relative; top:0px; left:- | + | #HumanPractice_Sublist{position:relative; top:0px; left:-470px;} |
#iGEM_logo{position:absolute; top:30px; left:1090px; height:80px;} | #iGEM_logo{position:absolute; top:30px; left:1090px; height:80px;} | ||
| Line 171: | Line 171: | ||
#AbstractBox{position:absolute; left:430px; top:390px; width:710px; height:540px; background-color:transparent;} | #AbstractBox{position:absolute; left:430px; top:390px; width:710px; height:540px; background-color:transparent;} | ||
#AbstractBackground{width:100%; height:100%; background-color:#FFFFFF; opacity:0.8;} | #AbstractBackground{width:100%; height:100%; background-color:#FFFFFF; opacity:0.8;} | ||
| - | #AbstractContent{position:absolute; left: | + | #AbstractContent{position:absolute; left:40px; top:25px; width:630px; font-size:18px; line-height:25px ; font-family: calibri, Arial, sans-serif; text-align:justify;} |
#AbstractContent:first-letter{font-size:30px;} | #AbstractContent:first-letter{font-size:30px;} | ||
.SmallBoxes{width:340px; height:250px;background-color:#FFFFFF; overflow:hidden;font-family: calibri, Arial, Helvetica, sans-serif; } | .SmallBoxes{width:340px; height:250px;background-color:#FFFFFF; overflow:hidden;font-family: calibri, Arial, Helvetica, sans-serif; } | ||
| Line 219: | Line 219: | ||
</li> | </li> | ||
<li id="PKU_navbar_Team" class="Navbar_Item"> | <li id="PKU_navbar_Team" class="Navbar_Item"> | ||
| - | <a >Team</a> | + | <a href="">Team</a> |
<ul id="Team_Sublist"> | <ul id="Team_Sublist"> | ||
<div class="BackgroundofSublist"></div> | <div class="BackgroundofSublist"></div> | ||
| Line 235: | Line 235: | ||
<li><a href="https://2013.igem.org/Team:Peking/Project/Plugins">Adaptors</a></li> | <li><a href="https://2013.igem.org/Team:Peking/Project/Plugins">Adaptors</a></li> | ||
<li><a href="https://2013.igem.org/Team:Peking/Project/BandpassFilter">Band-pass Filter</a></li> | <li><a href="https://2013.igem.org/Team:Peking/Project/BandpassFilter">Band-pass Filter</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Peking/Project/Devices">Devices</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
<li id="PKU_navbar_Model" class="Navbar_Item"> | <li id="PKU_navbar_Model" class="Navbar_Item"> | ||
| - | <a href=" | + | <a href="">Model</a> |
<ul id="Model_Sublist"> | <ul id="Model_Sublist"> | ||
| + | <div class="BackgroundofSublist"></div> | ||
| + | <li><a href="https://2013.igem.org/Team:Peking/Model">Band-pass Filter</a></li> | ||
| + | <li><a href="https://2013.igem.org/Team:Peking/ModelforFinetuning">Biosensor Fine-tuning</a></li> | ||
</ul> | </ul> | ||
</li> | </li> | ||
<li id="PKU_navbar_HumanPractice" class="Navbar_Item" style="width:90px"> | <li id="PKU_navbar_HumanPractice" class="Navbar_Item" style="width:90px"> | ||
| - | <a >Data page</a> | + | <a href="">Data page</a> |
<ul id="DataPage_Sublist"> | <ul id="DataPage_Sublist"> | ||
<div class="BackgroundofSublist"></div> | <div class="BackgroundofSublist"></div> | ||
| Line 260: | Line 264: | ||
<ul id="HumanPractice_Sublist"> | <ul id="HumanPractice_Sublist"> | ||
<div class="BackgroundofSublist"></div> | <div class="BackgroundofSublist"></div> | ||
| - | <li><a href="https://2013.igem.org/Team:Peking/HumanPractice/Questionnaire">Questionnaire</a></li> | + | <li><a href="https://2013.igem.org/Team:Peking/HumanPractice/Questionnaire">Questionnaire Survey</a></li> |
| - | <li><a href="https://2013.igem.org/Team:Peking/HumanPractice/FactoryVisit"> | + | <li><a href="https://2013.igem.org/Team:Peking/HumanPractice/FactoryVisit">Visit and Interview</a></li> |
<li><a href="https://2013.igem.org/Team:Peking/HumanPractice/ModeliGEM">Practical Analysis</a></li> | <li><a href="https://2013.igem.org/Team:Peking/HumanPractice/ModeliGEM">Practical Analysis</a></li> | ||
<li><a href="https://2013.igem.org/Team:Peking/HumanPractice/iGEMWorkshop">Team Communication</a></li> | <li><a href="https://2013.igem.org/Team:Peking/HumanPractice/iGEMWorkshop">Team Communication</a></li> | ||
| Line 269: | Line 273: | ||
</ul> | </ul> | ||
| - | <a href="https://igem.org/ | + | <a href="https://2013.igem.org/Main_Page"><img id="iGEM_logo" src="https://static.igem.org/mediawiki/igem.org/4/48/Peking_igemlogo.jpg"/></a> |
</div> | </div> | ||
<!--end navigationbar--> | <!--end navigationbar--> | ||
| Line 282: | Line 286: | ||
<p id="PreLocation"> | <p id="PreLocation"> | ||
| - | <B>★Presentation:</B> | + | <B>★Presentation:</B> Room 34-101, Saturday 12:30 PM, Session 2 |
| - |     <B>★Poster:</B> | + |     <B>★Poster:</B> Sunday, #13 (Stata) |
</p> | </p> | ||
| Line 342: | Line 346: | ||
</div> | </div> | ||
<div id="AppendixBox5" class="SmallBoxes"> | <div id="AppendixBox5" class="SmallBoxes"> | ||
| - | <h1 id="Appendix5Title"><i>Communications, Questionnaire, | + | <h1 id="Appendix5Title"><i>Communications, Questionnaire survey, <br/>Factory Visit</i> and interview</h1> |
<div id="Appendix5Cover" > | <div id="Appendix5Cover" > | ||
<img id="AppendixImg5" src="https://static.igem.org/mediawiki/igem.org/d/da/Peking2013_home_appendix5.jpg" /> | <img id="AppendixImg5" src="https://static.igem.org/mediawiki/igem.org/d/da/Peking2013_home_appendix5.jpg" /> | ||
<h1 class="AppendixHeadLine">Human Practice</h1> | <h1 class="AppendixHeadLine">Human Practice</h1> | ||
<img id="Appendix5Icon" class="AppendixIcon" src="https://static.igem.org/mediawiki/igem.org/8/85/Peking2013_home_appendix5icon.png" /> | <img id="Appendix5Icon" class="AppendixIcon" src="https://static.igem.org/mediawiki/igem.org/8/85/Peking2013_home_appendix5icon.png" /> | ||
| - | <p>To obtain the information about public awareness and the situation of aromatics pollution, a survey including factory | + | <p>To obtain the information about public awareness and the situation of aromatics pollution, a survey including factory visit and questionnaires has been conducted. We also guided an iGEM HS team and held "Model iGEM" as a competition without competitiveness.</p> |
<a href="https://2013.igem.org/Team:Peking/HumanPractice">LEARN MORE</a> | <a href="https://2013.igem.org/Team:Peking/HumanPractice">LEARN MORE</a> | ||
</div> | </div> | ||
Latest revision as of 18:10, 28 October 2013

Aromatics Scouts
A Comprehensive Biosensor Toolkit for Profiling Aromatic Compounds in the Environment
★Presentation: Room 34-101, Saturday 12:30 PM, Session 2 ★Poster: Sunday, #13 (Stata)

Monitoring aromatic compounds in the environment remains a substantial challenge today. Noting the power of biosensors for quick and convenient testing, Peking iGEM has developed a functionally comprehensive biosensor toolkit to profile aromatics in the environment.
Transcriptional regulators that each detect a specific class of aromatics were first bioinformatically determined; and then utilized to build a comprehensive set of biosensor circuits. Characterization on the detection profiles of individual biosensors and the orthogonality/crosstalk between them proved that these biosensors are very capable at profiling aromatics present in water.
Moreover, for the ease of practical applications, two types of genetic devices were also developed as plug-ins for biosensors: "Adaptors", a set of conceptually novel devices to convert undetectable compounds into detectable compounds, and "Band-pass Filter", a "concentration filter" that allows the detection of analyte concentration within a specific range.
We expect that these novel biosensors, together with the plug-in devices, will serve as intriguing synthetic biological tools for diverse practical applications.
Mining aromatics-sensing Biobricks
from the genomic database

Biosensor Mining
The core component of our biosensor toolkit is the transcriptional regulators that sense aromatics. For the comprehensiveness of aromatics-sensing, a data-mining process was conducted to mine transcriptional regulators for each typical class of aromatics from the database Uniprot.
LEARN MOREHigh-performance, profile-
specific biosensors

Biosensors
A comprehensive set of biosensor circuits have been implemented using the aromatics-sensing transcriptional regulators. Each biosensor has a specific aromatics-sensing profile. Furthermore, the orthogonality of their sensing profiles was carefully assessed for practical applications.
LEARN MOREConverting the undetectable into
the detectable

Adaptors
To expand the detection profiles of some biosensors, aromatics-metabolizing enzymes were taken from natural metabolic pathways, working as Adaptors to convert undetectable chemicals into detectable aromatics when coupled with biosensor circuits.
LEARN MORERapidly determining
the analyte concentration
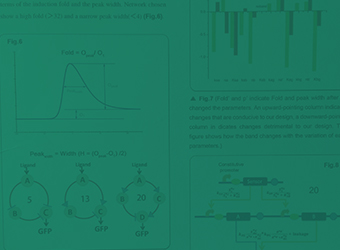
Band-pass Filter
For the ease of practical analysis, a genetic device called "Band-pass Filter" has been constructed to allow the detection of analyte concentration within a specific range. Biosensors equipped with the Band-pass Filter can robustly quantify the aromatics in environmental samples.
LEARN MORECommunications, Questionnaire survey,
Factory Visit and interview

Human Practice
To obtain the information about public awareness and the situation of aromatics pollution, a survey including factory visit and questionnaires has been conducted. We also guided an iGEM HS team and held "Model iGEM" as a competition without competitiveness.
LEARN MORE
 "
"

