Team:Newcastle/Parts/l form switch
From 2013.igem.org
| (186 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Newcastle}} | {{Team:Newcastle}} | ||
| - | =Switch | + | =Rod to L-form Switch BioBrick= |
| - | ==Purpose and | + | ==Purpose and Justification== |
| - | |||
| - | This | + | The purpose of this main ‘keystone’ BioBrick is to facilitate the switching of ''Bacillus subtilis'' cells from rod-shape to L-form, wall-less cells. Furthermore, this ‘switch’ enables L-form cells to return to rod-shape when required. This is facilitated through the introduction of a xylose-controlled promoter (''PxylR'') upstream of the ''murE'' gene, which is involved in the biosynthesis of peptidoglycan. The presence, or absence of a cell wall can be controlled through the presence, or absence of xylose, respectively. The antibiotic resistance marker chloramphenicol acetyl-transferase (''cat'') is upstream of this promoter region to allow for the selection of cells in which this system is integrated. |
| - | + | Homologous recombination allows this BioBrick to be integrated into the chromosome of ''B. subtilis''. The ''cat'' and ''PxylR'' sequences are flanked by sequences that are homologous with regions of the ''B. subtilis'' chromosome, which allows for insertion of the BioBrick at the intended loci. ~300bp at the 5’ end of the BioBrick is homologous with the end of the ''pbpB'' gene. Similarly ~300bp at the 3’ end of the biobrick is homologous with the start of the ''murE'' gene. | |
| - | + | Integration of the BioBrick facilitates xylose-mediated control over the expression of ''murE''. This cascades through biosynthetic pathways to enable control over peptidoglycan synthesis and thus control over cell wall production. Simply, in ''B. subtilis'' cells that have integrated the BioBrick into their chromosome, only in the presence of xylose is the cell wall produced. When xylose is not present cells lose their cell wall and survivors adopt the L-form phenotype. | |
| - | + | The model of the L-form BioBrick [http://parts.igem.org/Part:BBa_K1185000 BBa_K1185000] can be found on the Registry of Standard Biological Parts. | |
| - | + | <!--https://static.igem.org/mediawiki/2013/3/33/BareCillus_Switch.png--> | |
| - | + | ||
| - | + | [[File:BareCillus_Switch.png|700px]] | |
| - | + | ||
| - | + | ==Design== | |
| - | + | ||
| - | + | [[File:Switch 1.png|700px]] | |
| - | + | ''Figure 1. Showing all of the components of L-form switch BioBrick starting from the pbpb homology region at the far left, Spacer sequence, Chloramphenicol resistance marker, another Spacer, Pxyl promoter, murE RBS and finally the murE homology region at the far right.'' | |
| + | <br> | ||
| - | + | The L-form switch BioBrick is based upon the regulatory sequence controlling expression of ''murE'' in ''B. subtilis'' LR2 - a strain capable of L-form state. In this ''B. subtilis'' strain the peptidoglycan synthesis pathway required for the cell wall can be disrupted through control of ''murE'' expression - responsible for the synthesis of enzymes involved in peptidoglycan precursor biosynthesis. A xylose-contolled ''PxylR'' promoter upstream of ''murE'' facilitates this control. Upstream of this promoter the LR2 strain also contains a ''cat'' gene - conferring chloramphenicol resistance. These genetic components differentiate the ''B. subtilis'' LR2 chromosome from that of ''B. subtilis'' 168. On the LR2 chromosome these genetic components take the place of the ''spoVD'' gene (involved in sporulation) in ''B. subtilis'' 168. It is these genetic elements that have been utilised to create a BioBrick that facilitates the switching of ''B. subtilis'' cells from rod state to L-form, and back again. The flanking regions of the ''cat'' gene and the ''PxylR'' promoter - part of the ''pbpB'' coding sequence at the 5' end of this region of DNA and part of the ''murE'' coding sequence at the 3' end - are also of vital importance, allowing for integration of the BioBrick into the chromosome of ''B. subtilis'' via homologous recombination. | |
| - | |||
| - | + | [[File:Switch.png|700px]] | |
| - | + | ''Figure 2. Diagram showing how the L-form switch BioBrick is integrated into the B. subtilis chromosome.'' | |
| - | + | In order to package the genetic elements within the ''B. subtilis'' chromosome that enable this strain to transition from rod to L-form, the full sequence of the region between the ''pbpB'' and the ''murE'' genes had to be identified. The sequence for the L-form switch region was obtained through a de novo assembly (using Sequencher 5.1 DNA sequencing software) of sequence data gained from sequencing between the ''pbpB'' and ''murE'' genes on the ''B. subtilis'' LR2 chromosome (using designed primers) with reads produced by high throughput sequencing of the full ''B. subtilis'' LR2 genome. In particular, reads that did not assemble when using ''B. subtilis'' 168 as a reference genome were used in this assembly. The primers that were designed to sequence the region between the ''pbpB'' and ''murE'' genes on the ''B. subtilis'' LR2 chromosome allowed sequencing from both the 5’ and the 3' ends of the desired sequence. The design of the primers allowed for sequencing to include the final 309bp at the 3’ end of the ''pbpB'' coding sequence (upstream of the L-form switch region), as well as the sequence of the 331bp at the 5’ end of the ''murE'' coding sequence (downstream, and under the control, of the L-form switch). Once fully sequenced, this region of DNA was able to be synthesised once compliant with the [http://parts.igem.org/Assembly_standard_10 BioBrick RFC[10]] standard. | |
| - | + | To allow the L-form switch BioBrick to be submitted to the iGEM registry of standard biological parts it needed to comply with BioBrick RFC[10] standard. In order to comply with this standard the BioBrick RFC[10] prefix and suffix sequences were added to the synthetic construct sequence using Sequencher 5.1 DNA sequencing software. Two EcoRI restriction sites and a PstI restriction site that were present in the sequence were removed through single nucleotide changes using Gene Designer 2.0 - these restriction sites violated the terms of the RFC[10] standard. Base changes were chosen to optimise the altered codons (in the open reading frame of coding sequences) for codon bias in ''B. subtilis''. The altered genetic sequence was then able to be synthesised. | |
| - | + | ==Modelling== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | To predict if the BioBrick we designed would function as desired, we modelled [https://2013.igem.org/Team:Newcastle/Modelling/L-form_Switch peptidoglycan biosynthesis] as peptidoglycan is the major component of the cell wall. The model suggested that peptidoglycan would not be synthesised if ''murE'' was switched off. | |
| - | |||
| - | + | ==Construction== | |
| - | + | The designed BioBrick was synthesised by DNA synthesis company DNA 2.0. The components within the BioBrick were as follows: - | |
| - | + | Parts: | |
| + | #Biobrick prefix – RFC[10] standard. | ||
| + | #309bp of the end of the ''pbpB'' coding sequence. | ||
| + | #Spacer. | ||
| + | #Reverse and complement of chloramphenicol acetyl-transferase (''cat'') coding sequence (including native ribosome binding site (RBS) and promoter). | ||
| + | #''PxylR'' promoter. | ||
| + | #Spacer. | ||
| + | #RBS binding site for ''murE''. | ||
| + | #Spacer. | ||
| + | #331bp of the start of ''murE'' coding sequence. | ||
| + | #Biobrick suffix – RFC[10] standard. | ||
| - | + | The synthesised BioBrick was then inserted into the pSB1C3 plasmid - the only plasmid acceptable by iGEM for part submission. | |
| - | + | ==Cloning and Integration== | |
| - | 2. | + | After the pSB1C3 plasmid containing the L-form switch BioBrick was returned from DNA 2.0, it was amplified in ''E.coli''. The [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Escherichia_coli_Competent_Cell_Preparation ''E. coli'' competent cell preparation] and [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Escherichia_coli_Transformation ''E. coli transformation''] protocols were used to transform this plasmid into ''E. coli'' cells for cloning. Transformant cells were incubated in order to clone the transformed plasmids. Then the cloned plasmids were [https://2013.igem.org/Team:Newcastle/Notebook/protocols#QIAprep_Spin_Miniprep_Kit extracted] and [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Bacillus_subtilis_168_Competent_cell_Prep_and_Transformation transformed] into ''B. subtilis''. Integration of the BioBrick from the vector plasmid to the host chromosome was facilitated through homologous recombination with crossover events within the ''pbpB'' and ''murE'' genes. |
| - | + | ||
| - | + | ||
| - | + | [[File:Switch.png|centre|700px]] | |
| - | + | ''Figure 2. Diagram showing how the L-form switch BioBrick is integrated into the B. subtilis chromosome.'' | |
| - | + | <!--[[File:L-form biobrick construct.png|400px]]--> | |
| - | == | + | ==Testing and Characterisation== |
| - | + | In order to produce L-forms we transformed the switch BioBrick into ''Bacillus subtilis'' str.168 and inoculated the transformants into LB media containing 5ug/ml Chloramphenicol and 0.8% Xylose. As can be seen on Figure 3, colonies grew on both plate 1 (''B. Subtilis'' str. 168 + (Switch BioBrick)) and plate 2 (positive control, ''B.subtilis'' str. 168 + pGFPrrnB) as plasmid pMutin4 confers chloramphenicol resistance to ''B. subtilis''. There were no colonies growing on Plate 3 which was inoculated with ''B. subtilis'' str. 168 into the same media (negative control) as ''B. subtilis'' str. 168 does not naturally have resistance to chloramphenicol. These transformation results suggest that the switch BioBrick was transformed into ''B. subtilis'' str.168 successfully. | |
| - | + | <html> | |
| + | <style type="text/css"> | ||
| + | #body img{ | ||
| + | margin-bottom:3px; | ||
| + | } | ||
| + | table{ | ||
| + | border:2px solid black; | ||
| + | margin-left:50px; | ||
| + | margin-bottom: 10px; | ||
| + | width: 650px; | ||
| + | } | ||
| + | .italic{ | ||
| + | font-style: italic; | ||
| + | } | ||
| + | table.borderless { | ||
| + | border:0px solid white; | ||
| + | } | ||
| - | 3. | + | </style> |
| + | <body> | ||
| + | <br> | ||
| + | <table border="0"> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/d/df/BareCillus_168%2BSwitch.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 1: B. Subtilis str. 168 transformed with switch BioBrick.</div></td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/5/54/BareCillus_168%2BrrnB.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 2: B.subtilis str. 168 transformed with pGFPrrnB (positive control).</div></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/6/62/BareCillus_168%2BCm%2BH20.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 3: B. subtilis str. 168 no transformation (negative control). | ||
| + | </div></td> | ||
| + | <td><div class="italic">Figure 3. Plates of B. Subtilis str. 168 transformed with switch BioBrick, pGFPrrnB (positive control) and water (negative control).</div></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </body> | ||
| + | </html> | ||
| - | + | Even though colonies were found on Plate 1, we couldn't conclude that our BioBrick had integrated into the ''pbpb'' and ''murE'' homology region replacing the ''spoVD'' section with the chloramphenicol resistance cassete and the Pxyl promoter. So, in order to test that the transformants had actually taken up the switch BioBrick and integrated it into the homology region in the ''B. subtilis'' chromosomes, some colonies were picked from Plate 1 and inoculated into NB/MSM media with xylose concentration varying between 0% and 0.8% and incubated in 30oC for 56 hours. After this incubation the morphology of the cells was checked under the microscope (Figure 4). In the batch of ''B. Subtilis'' incubated without xylose, all of the cells appeared as L-forms and in 0.2% xylose the majority of the cells were L-forms with only occasional rods. In 0.4% and 0.5% xylose the majority of the cells were rods with only a couple of L-forms being spotted and 0.6% and 0.8% xylose all cells appeared as rods. The results of this experiment have been summarised into table and graph formats which is displayed on Table 1 and Graph 1. These results allow us to conclude that our switch BioBrick works as designed and as the model suggested it would. | |
| - | + | ||
| - | === | + | <html> |
| - | + | <body> | |
| + | <div id="tableWrap"> | ||
| + | <table border="0"> | ||
| + | <tr> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/3/38/BareCillus_0.jpg" alt="Pulpit rock" width="268px" height="208px"><br><i>O% xylose</i></td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/a/a6/BareCillus_0.2.jpg" alt="Pulpit rock" width="268px" height="208px"><br>0.2% xylose</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/6/67/BareCillus_0.4.jpg" alt="Pulpit rock" width="268px" height="208px"><br>0.4% xylose</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/c/cd/BareCillus_0.5.jpg" alt="Pulpit rock" width="268px" height="208px"><br>0.5% xylose | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/9/91/BareCillus_0.6.jpg" alt="Pulpit rock" width="268px" height="208px"><br>0.6% xylose</td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/2/25/BareCillus_0.8.jpg" alt="Pulpit rock" width="268px" height="208px"><br>0.8% xylose</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | </body> | ||
| + | </html> | ||
| - | + | ''Figure 4. This shows the appearance of B. subtilis cells transformed with our switch BioBrick after being inoculated into a 1:1 ration of NB and MSM media with a range of xylose concentrations and incubated in 30oC for 56 hours. The xylose concentrations in %(w/v) are denoted under each image.'' | |
| - | + | <table class="data"> | |
| + | <tr> | ||
| + | <th>%(w/v)xylose</th> | ||
| + | <th>Number of rods</th> | ||
| + | <th>Number of L-forms</th> | ||
| + | <th>Total cell number</th> | ||
| + | <th>% L-forms</th> | ||
| + | <th>% Rods</th> | ||
| + | </tr> | ||
| + | <tr class="odd"> | ||
| + | <td class="data">0</td> | ||
| + | <td class="data">0</td> | ||
| + | <td class="data">16</td> | ||
| + | <td class="data">16</td> | ||
| + | <td class="data">100</td> | ||
| + | <td class="data">0</td> | ||
| + | </tr> | ||
| + | <tr class="even"> | ||
| + | <td class="data">0.2</td> | ||
| + | <td class="data">1</td> | ||
| + | <td class="data">21</td> | ||
| + | <td class="data">22</td> | ||
| + | <td class="data">95</td> | ||
| + | <td class="data">5</td> | ||
| + | </tr> | ||
| + | <tr class="odd"> | ||
| + | <td class="data">0.4</td> | ||
| + | <td class="data">26</td> | ||
| + | <td class="data">10</td> | ||
| + | <td class="data">36</td> | ||
| + | <td class="data">28</td> | ||
| + | <td class="data">72</td> | ||
| + | </tr> | ||
| + | <tr class="even"> | ||
| + | <td class="data">0.5</td> | ||
| + | <td class="data">207</td> | ||
| + | <td class="data">3</td> | ||
| + | <td class="data">10</td> | ||
| + | <td class="data">1</td> | ||
| + | <td class="data">99</td> | ||
| - | === | + | </tr> |
| - | + | <tr class="odd"> | |
| - | + | <td class="data">0.6</td> | |
| + | <td class="data">450</td> | ||
| + | <td class="data">0</td> | ||
| + | <td class="data">450</td> | ||
| + | <td class="data">0</td> | ||
| + | <td class="data">100</td> | ||
| + | </tr> | ||
| + | <tr class="even"> | ||
| + | <td class="data">0.8</td> | ||
| + | <td class="data">480</td> | ||
| + | <td class="data">0</td> | ||
| + | <td class="data">480</td> | ||
| + | <td class="data">0</td> | ||
| + | <td class="data">100</td> | ||
| - | + | </tr> | |
| + | |||
| + | </table> | ||
| - | + | ''Table 1. Number of L-forms and rod cells and ratio of L-forms to rod cells of B. Subtilis 168 containing our switch BioBrick in Different xylose concentrations after 56 hours Incubation '' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[File:Xylose conc.jpg|700px]] | |
| + | ''Graph 1. Ratio of L-forms to rod cells of B. Subtilis 168 containing our switch BioBrick in Different xylose concentrations after 56 hours Incubation'' | ||
| - | + | We also considered using Flow cytometry to sort between L-forms and rods cells over a period of 72 hours to give us more detailed and accurate results regarding when cell actually starts to turn into L-forms from rods in media with different xylose concentrations. However, due to the high viscosity of the media (NB/MSM) which contains sucrose that L-forms need to survive, we couldn't calibrate the Flow Cytometry to do the reading. | |
| - | + | To further test the switch BioBrick, we also transformed it into ''B. subtilis'' BSB1. As can be seen on Figure 5, there were colonies growing on both Plate 4 (''B. Subtilis'' BSB1 + (Switch BioBrick)) and also on Plate 5 (positive control, ''B.subtilis'' BSB1 + pGFPrrnB) as plasmid pMutin4 confers chloramphenicol resistance. There were no colonies growing on Plate 6 which was inoculated with ''B. subtilis'' BSB1 onto the same media (negative control)as ''B. subtilis'' BSB1 does not naturally have resistance to chloramphenicol. With these results, we can conclude that the switch BioBrick can be successfully transformed and integrated into both commonly used ''B. subtilis'' lab strains. | |
| - | + | <html> | |
| + | <style type="text/css"> | ||
| + | #body img{ | ||
| + | margin-bottom:3px; | ||
| + | } | ||
| + | table{ | ||
| + | border:2px solid black; | ||
| + | margin-left:50px; | ||
| + | margin-bottom: 10px; | ||
| + | width: 650px; | ||
| + | } | ||
| + | .italic{ | ||
| + | font-style: italic; | ||
| + | } | ||
| + | table.borderless { | ||
| + | border:0px solid white; | ||
| + | } | ||
| - | + | </style> | |
| + | <body> | ||
| + | <br> | ||
| + | <table border="0"> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/3/3f/BareCillus_BSB1%2BSwitch.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 4: B. Subtilis BSB1 transformed with the switch BioBrick.</div></td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/1/1f/BareCillus_BSB1%2Brrnb.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 5: B. Subtilis BSB1 transformed with the pGFPrrnB (positive control).</div></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/a/ae/BareCillus_BSB1%2BCm%2BH20.jpg" alt="Pulpit rock" width="304" height="228"><div class="italic">Plate 6: B. Subtilis BSB1, not transformed (negative control). | ||
| + | </div></td> | ||
| + | <td><div class="italic">Figure 5. Plates of B. Subtilis str. BSB1 transformed with switch BioBrick, pGFPrrnB (positive control) and water (negative control).</div></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </body> | ||
| + | </html> | ||
| - | + | The images in figure 6 and 7 show the different morphology of ''B. subtilis'' str. 168 and BSB1 before the integration of the switch BioBrick and following 56 hours incubation in 0% Xylose NB/MSM media. | |
| - | |||
| - | |||
| - | + | <html> | |
| + | <body> | ||
| + | <style type="text/css"> | ||
| + | table{ | ||
| + | border:2px solid black; | ||
| + | margin-left:50px; | ||
| + | margin-bottom: 10px; | ||
| + | width: 650px; | ||
| + | } | ||
| + | #tableWrap{ | ||
| + | font-style: italic; | ||
| + | } | ||
| + | </style> | ||
| + | <table border="0"> | ||
| + | <tr> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/8/83/168_rod.jpg" alt="Pulpit rock" width="268px" height="208px"><br><i>Figure 6. B. subtilis str. 168 prior to transformation with our switch BioBrick</i></td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/0/0e/168_L-form.jpg" alt="Pulpit rock" width="268px" height="208px"><br><i>B. subtilis str. 168 after to transformation with our switch BioBrick</i></td> | ||
| + | </tr> | ||
| + | </table> | ||
| - | + | <table border="0"> | |
| + | <tr> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/2/2c/BSB1_rod.jpg" alt="Pulpit rock" width="268px" height="208px"><br><i>Figure 7. B. subtilis str. BSB1 prior to transformation with our switch BioBrick</i></td> | ||
| + | <td><img src="https://static.igem.org/mediawiki/2013/4/45/BSB1_L-form.jpg" alt="Pulpit rock" width="268px" height="208px"><br><i>B. subtilis str. BSB1 after to transformation with our switch BioBrick</i></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </body> | ||
| + | </html> | ||
| - | + | This method of generating L-forms is very time consuming, and follows the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#L-form_Growth_Media Liquid media protocol]. Figure 8 and Video 1 show L-forms being generated from ''B. subtilis'' rods, a process taking 12 hours. The rod-shaped cells can be seen shortening and widening over time as peptidoglycan cell wall synthesis is down-regulated and there is insufficient amounts of cell wall to maintain the regular rod-form cell shape. This results in the eventual adoption of circular L-form shape (visible at 686.25 minutes in Figure 8). Cells that achieve this form are completely unbound by cell wall. | |
| - | + | ||
| - | + | In order to speed things up you can use lysozyme to initially remove the cell wall as in our [https://2013.igem.org/Team:Newcastle/Notebook/protocols#Protoplasting_to_generate_L-form Lysozyme protoplasting protocol]. The L-forms, as explained in the L-form page, will still be able to grow and divide, as opposed to regular protoplasts. | |
| - | + | [[File:Rod-L.jpg|600px]] | |
| - | + | ''Figure 8. Fluorescence microscopy showing B. subtilis rod cells turning into L-forms over a period of nearly 12 hours following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#L-form_Growth_Media Liquid media protocol.] '' | |
| - | |||
| - | + | <html> | |
| + | <style type="text/css"> | ||
| + | #video { | ||
| + | margin-left: 50px; | ||
| + | } | ||
| + | </style> | ||
| + | <div id="video"> | ||
| + | <video src="https://static.igem.org/mediawiki/2013/d/de/Rod_to_L_form_conversion.mp4" width="640" height="360" controls preload></video> | ||
| + | </div> | ||
| + | </html> | ||
| - | + | ''Video 1. Time-lapse showing the conversion of B. subtilis rod cells to L-forms over a period of nearly 12 hours following the [https://2013.igem.org/Team:Newcastle/Notebook/protocols#L-form_Growth_Media Liquid media protocol.] '' | |
| - | + | ==References== | |
| - | + | [http://www.ncbi.nlm.nih.gov/pubmed/19212404 Leaver M., Dominguez-CuevasP., Coxhead J.M., Daniel R.A. and Errington J. (2009) Life without a wall or division machine in Bacillus subtilis. ''Nature'', '''457''', 849-853.] | |
| - | + | [http://www.ncbi.nlm.nih.gov/pubmed/23452849 Mercier R., Kawai Y. and Errington J. (2013) Excess membrane synthesis drives a primitive mode of cell proliferation. ''Cell'', '''152''', 997-1007.] | |
| - | + | {{Team:Newcastle/Sponsors}} | |
Latest revision as of 18:35, 28 October 2013

Contents |
Rod to L-form Switch BioBrick
Purpose and Justification
The purpose of this main ‘keystone’ BioBrick is to facilitate the switching of Bacillus subtilis cells from rod-shape to L-form, wall-less cells. Furthermore, this ‘switch’ enables L-form cells to return to rod-shape when required. This is facilitated through the introduction of a xylose-controlled promoter (PxylR) upstream of the murE gene, which is involved in the biosynthesis of peptidoglycan. The presence, or absence of a cell wall can be controlled through the presence, or absence of xylose, respectively. The antibiotic resistance marker chloramphenicol acetyl-transferase (cat) is upstream of this promoter region to allow for the selection of cells in which this system is integrated.
Homologous recombination allows this BioBrick to be integrated into the chromosome of B. subtilis. The cat and PxylR sequences are flanked by sequences that are homologous with regions of the B. subtilis chromosome, which allows for insertion of the BioBrick at the intended loci. ~300bp at the 5’ end of the BioBrick is homologous with the end of the pbpB gene. Similarly ~300bp at the 3’ end of the biobrick is homologous with the start of the murE gene.
Integration of the BioBrick facilitates xylose-mediated control over the expression of murE. This cascades through biosynthetic pathways to enable control over peptidoglycan synthesis and thus control over cell wall production. Simply, in B. subtilis cells that have integrated the BioBrick into their chromosome, only in the presence of xylose is the cell wall produced. When xylose is not present cells lose their cell wall and survivors adopt the L-form phenotype.
The model of the L-form BioBrick [http://parts.igem.org/Part:BBa_K1185000 BBa_K1185000] can be found on the Registry of Standard Biological Parts.
Design
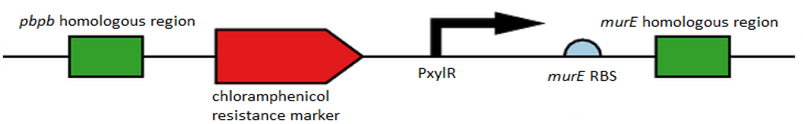
Figure 1. Showing all of the components of L-form switch BioBrick starting from the pbpb homology region at the far left, Spacer sequence, Chloramphenicol resistance marker, another Spacer, Pxyl promoter, murE RBS and finally the murE homology region at the far right.
The L-form switch BioBrick is based upon the regulatory sequence controlling expression of murE in B. subtilis LR2 - a strain capable of L-form state. In this B. subtilis strain the peptidoglycan synthesis pathway required for the cell wall can be disrupted through control of murE expression - responsible for the synthesis of enzymes involved in peptidoglycan precursor biosynthesis. A xylose-contolled PxylR promoter upstream of murE facilitates this control. Upstream of this promoter the LR2 strain also contains a cat gene - conferring chloramphenicol resistance. These genetic components differentiate the B. subtilis LR2 chromosome from that of B. subtilis 168. On the LR2 chromosome these genetic components take the place of the spoVD gene (involved in sporulation) in B. subtilis 168. It is these genetic elements that have been utilised to create a BioBrick that facilitates the switching of B. subtilis cells from rod state to L-form, and back again. The flanking regions of the cat gene and the PxylR promoter - part of the pbpB coding sequence at the 5' end of this region of DNA and part of the murE coding sequence at the 3' end - are also of vital importance, allowing for integration of the BioBrick into the chromosome of B. subtilis via homologous recombination.
Figure 2. Diagram showing how the L-form switch BioBrick is integrated into the B. subtilis chromosome.
In order to package the genetic elements within the B. subtilis chromosome that enable this strain to transition from rod to L-form, the full sequence of the region between the pbpB and the murE genes had to be identified. The sequence for the L-form switch region was obtained through a de novo assembly (using Sequencher 5.1 DNA sequencing software) of sequence data gained from sequencing between the pbpB and murE genes on the B. subtilis LR2 chromosome (using designed primers) with reads produced by high throughput sequencing of the full B. subtilis LR2 genome. In particular, reads that did not assemble when using B. subtilis 168 as a reference genome were used in this assembly. The primers that were designed to sequence the region between the pbpB and murE genes on the B. subtilis LR2 chromosome allowed sequencing from both the 5’ and the 3' ends of the desired sequence. The design of the primers allowed for sequencing to include the final 309bp at the 3’ end of the pbpB coding sequence (upstream of the L-form switch region), as well as the sequence of the 331bp at the 5’ end of the murE coding sequence (downstream, and under the control, of the L-form switch). Once fully sequenced, this region of DNA was able to be synthesised once compliant with the [http://parts.igem.org/Assembly_standard_10 BioBrick RFC[10]] standard.
To allow the L-form switch BioBrick to be submitted to the iGEM registry of standard biological parts it needed to comply with BioBrick RFC[10] standard. In order to comply with this standard the BioBrick RFC[10] prefix and suffix sequences were added to the synthetic construct sequence using Sequencher 5.1 DNA sequencing software. Two EcoRI restriction sites and a PstI restriction site that were present in the sequence were removed through single nucleotide changes using Gene Designer 2.0 - these restriction sites violated the terms of the RFC[10] standard. Base changes were chosen to optimise the altered codons (in the open reading frame of coding sequences) for codon bias in B. subtilis. The altered genetic sequence was then able to be synthesised.
Modelling
To predict if the BioBrick we designed would function as desired, we modelled peptidoglycan biosynthesis as peptidoglycan is the major component of the cell wall. The model suggested that peptidoglycan would not be synthesised if murE was switched off.
Construction
The designed BioBrick was synthesised by DNA synthesis company DNA 2.0. The components within the BioBrick were as follows: -
Parts:
- Biobrick prefix – RFC[10] standard.
- 309bp of the end of the pbpB coding sequence.
- Spacer.
- Reverse and complement of chloramphenicol acetyl-transferase (cat) coding sequence (including native ribosome binding site (RBS) and promoter).
- PxylR promoter.
- Spacer.
- RBS binding site for murE.
- Spacer.
- 331bp of the start of murE coding sequence.
- Biobrick suffix – RFC[10] standard.
The synthesised BioBrick was then inserted into the pSB1C3 plasmid - the only plasmid acceptable by iGEM for part submission.
Cloning and Integration
After the pSB1C3 plasmid containing the L-form switch BioBrick was returned from DNA 2.0, it was amplified in E.coli. The E. coli competent cell preparation and E. coli transformation protocols were used to transform this plasmid into E. coli cells for cloning. Transformant cells were incubated in order to clone the transformed plasmids. Then the cloned plasmids were extracted and transformed into B. subtilis. Integration of the BioBrick from the vector plasmid to the host chromosome was facilitated through homologous recombination with crossover events within the pbpB and murE genes.
Figure 2. Diagram showing how the L-form switch BioBrick is integrated into the B. subtilis chromosome.
Testing and Characterisation
In order to produce L-forms we transformed the switch BioBrick into Bacillus subtilis str.168 and inoculated the transformants into LB media containing 5ug/ml Chloramphenicol and 0.8% Xylose. As can be seen on Figure 3, colonies grew on both plate 1 (B. Subtilis str. 168 + (Switch BioBrick)) and plate 2 (positive control, B.subtilis str. 168 + pGFPrrnB) as plasmid pMutin4 confers chloramphenicol resistance to B. subtilis. There were no colonies growing on Plate 3 which was inoculated with B. subtilis str. 168 into the same media (negative control) as B. subtilis str. 168 does not naturally have resistance to chloramphenicol. These transformation results suggest that the switch BioBrick was transformed into B. subtilis str.168 successfully.
 Plate 1: B. Subtilis str. 168 transformed with switch BioBrick. |
 Plate 2: B.subtilis str. 168 transformed with pGFPrrnB (positive control). |
 Plate 3: B. subtilis str. 168 no transformation (negative control).
|
Figure 3. Plates of B. Subtilis str. 168 transformed with switch BioBrick, pGFPrrnB (positive control) and water (negative control). |
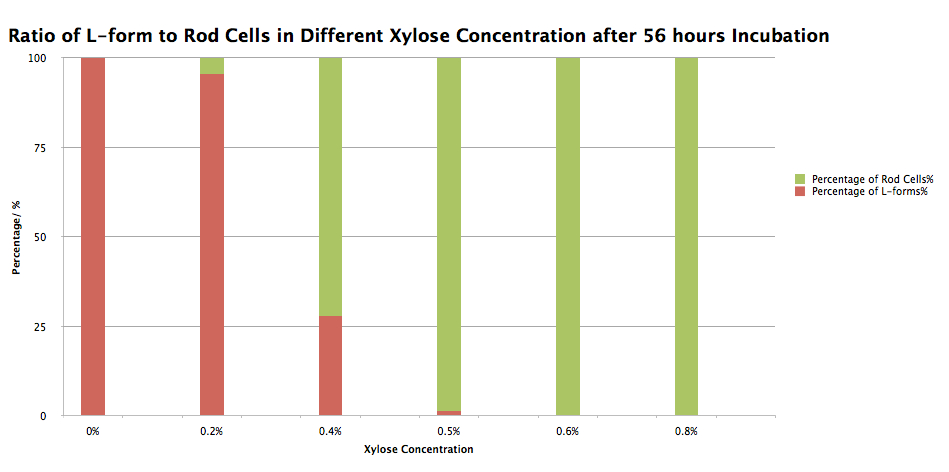
Even though colonies were found on Plate 1, we couldn't conclude that our BioBrick had integrated into the pbpb and murE homology region replacing the spoVD section with the chloramphenicol resistance cassete and the Pxyl promoter. So, in order to test that the transformants had actually taken up the switch BioBrick and integrated it into the homology region in the B. subtilis chromosomes, some colonies were picked from Plate 1 and inoculated into NB/MSM media with xylose concentration varying between 0% and 0.8% and incubated in 30oC for 56 hours. After this incubation the morphology of the cells was checked under the microscope (Figure 4). In the batch of B. Subtilis incubated without xylose, all of the cells appeared as L-forms and in 0.2% xylose the majority of the cells were L-forms with only occasional rods. In 0.4% and 0.5% xylose the majority of the cells were rods with only a couple of L-forms being spotted and 0.6% and 0.8% xylose all cells appeared as rods. The results of this experiment have been summarised into table and graph formats which is displayed on Table 1 and Graph 1. These results allow us to conclude that our switch BioBrick works as designed and as the model suggested it would.
 O% xylose |
 0.2% xylose |
 0.4% xylose |
 0.5% xylose |
 0.6% xylose |
 0.8% xylose |
Figure 4. This shows the appearance of B. subtilis cells transformed with our switch BioBrick after being inoculated into a 1:1 ration of NB and MSM media with a range of xylose concentrations and incubated in 30oC for 56 hours. The xylose concentrations in %(w/v) are denoted under each image.
| %(w/v)xylose | Number of rods | Number of L-forms | Total cell number | % L-forms | % Rods |
|---|---|---|---|---|---|
| 0 | 0 | 16 | 16 | 100 | 0 |
| 0.2 | 1 | 21 | 22 | 95 | 5 |
| 0.4 | 26 | 10 | 36 | 28 | 72 |
| 0.5 | 207 | 3 | 10 | 1 | 99 |
| 0.6 | 450 | 0 | 450 | 0 | 100 |
| 0.8 | 480 | 0 | 480 | 0 | 100 |
Table 1. Number of L-forms and rod cells and ratio of L-forms to rod cells of B. Subtilis 168 containing our switch BioBrick in Different xylose concentrations after 56 hours Incubation
Graph 1. Ratio of L-forms to rod cells of B. Subtilis 168 containing our switch BioBrick in Different xylose concentrations after 56 hours Incubation
We also considered using Flow cytometry to sort between L-forms and rods cells over a period of 72 hours to give us more detailed and accurate results regarding when cell actually starts to turn into L-forms from rods in media with different xylose concentrations. However, due to the high viscosity of the media (NB/MSM) which contains sucrose that L-forms need to survive, we couldn't calibrate the Flow Cytometry to do the reading.
To further test the switch BioBrick, we also transformed it into B. subtilis BSB1. As can be seen on Figure 5, there were colonies growing on both Plate 4 (B. Subtilis BSB1 + (Switch BioBrick)) and also on Plate 5 (positive control, B.subtilis BSB1 + pGFPrrnB) as plasmid pMutin4 confers chloramphenicol resistance. There were no colonies growing on Plate 6 which was inoculated with B. subtilis BSB1 onto the same media (negative control)as B. subtilis BSB1 does not naturally have resistance to chloramphenicol. With these results, we can conclude that the switch BioBrick can be successfully transformed and integrated into both commonly used B. subtilis lab strains.
 Plate 4: B. Subtilis BSB1 transformed with the switch BioBrick. |
 Plate 5: B. Subtilis BSB1 transformed with the pGFPrrnB (positive control). |
 Plate 6: B. Subtilis BSB1, not transformed (negative control).
|
Figure 5. Plates of B. Subtilis str. BSB1 transformed with switch BioBrick, pGFPrrnB (positive control) and water (negative control). |
The images in figure 6 and 7 show the different morphology of B. subtilis str. 168 and BSB1 before the integration of the switch BioBrick and following 56 hours incubation in 0% Xylose NB/MSM media.
 Figure 6. B. subtilis str. 168 prior to transformation with our switch BioBrick |
 B. subtilis str. 168 after to transformation with our switch BioBrick |
 Figure 7. B. subtilis str. BSB1 prior to transformation with our switch BioBrick |
 B. subtilis str. BSB1 after to transformation with our switch BioBrick |
This method of generating L-forms is very time consuming, and follows the Liquid media protocol. Figure 8 and Video 1 show L-forms being generated from B. subtilis rods, a process taking 12 hours. The rod-shaped cells can be seen shortening and widening over time as peptidoglycan cell wall synthesis is down-regulated and there is insufficient amounts of cell wall to maintain the regular rod-form cell shape. This results in the eventual adoption of circular L-form shape (visible at 686.25 minutes in Figure 8). Cells that achieve this form are completely unbound by cell wall.
In order to speed things up you can use lysozyme to initially remove the cell wall as in our Lysozyme protoplasting protocol. The L-forms, as explained in the L-form page, will still be able to grow and divide, as opposed to regular protoplasts.
Figure 8. Fluorescence microscopy showing B. subtilis rod cells turning into L-forms over a period of nearly 12 hours following the Liquid media protocol.
Video 1. Time-lapse showing the conversion of B. subtilis rod cells to L-forms over a period of nearly 12 hours following the Liquid media protocol.
References
[http://www.ncbi.nlm.nih.gov/pubmed/19212404 Leaver M., Dominguez-CuevasP., Coxhead J.M., Daniel R.A. and Errington J. (2009) Life without a wall or division machine in Bacillus subtilis. Nature, 457, 849-853.]
[http://www.ncbi.nlm.nih.gov/pubmed/23452849 Mercier R., Kawai Y. and Errington J. (2013) Excess membrane synthesis drives a primitive mode of cell proliferation. Cell, 152, 997-1007.]
 "
"