Team:Imperial College/Growth Assays
From 2013.igem.org
| (82 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
This page includes all of our experimental growth, toxicity and sole carbon source assay data. | This page includes all of our experimental growth, toxicity and sole carbon source assay data. | ||
| - | + | <b><font size="3"> OD600 in some experiments are larger than 1, as they were converted from absorbance at 600nm. Thus they are considered accurate.</font size="3"></b> | |
<h2>Growth assays with different experimental media</h2> | <h2>Growth assays with different experimental media</h2> | ||
In additional to standard LB and minimal media, several novel experimental media were developed in order to characterise Biobricks within a mixed waste/landfill setting. These media were characterised through an examination of pH and through an array of growth assays with the project chassis, E.coli (MG1655). | In additional to standard LB and minimal media, several novel experimental media were developed in order to characterise Biobricks within a mixed waste/landfill setting. These media were characterised through an examination of pH and through an array of growth assays with the project chassis, E.coli (MG1655). | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| - | |[[File:LB_M9.png|thumbnail|left| | + | |[[File:LB_M9.png|thumbnail|left|450px|<b>Media characterisation</b>. E. coli strain MG1655 were transformed with a control plasmid and grown in different experimental media over a period of 5 hours. LB media, minimal media ([[Team:Imperial_College/Protocols#M9_minimal_and_supplemented_media|M9M]]), supplemented minimal media (M9S), as described [https://2013.igem.org/Team:Imperial_College/Protocols#M9_minimal_and_supplemented_media here] or waste conditioned media (WCM), which is made from sterile filtrated mixed waste, see [https://2013.igem.org/Team:Imperial_College/Protocols#Waste_Conditioned_Media_.28WCM.29 here]. OD600 measured, error bars are S.E.M., n=4. Figure made by Imperial College London 2013 iGEM.]] |
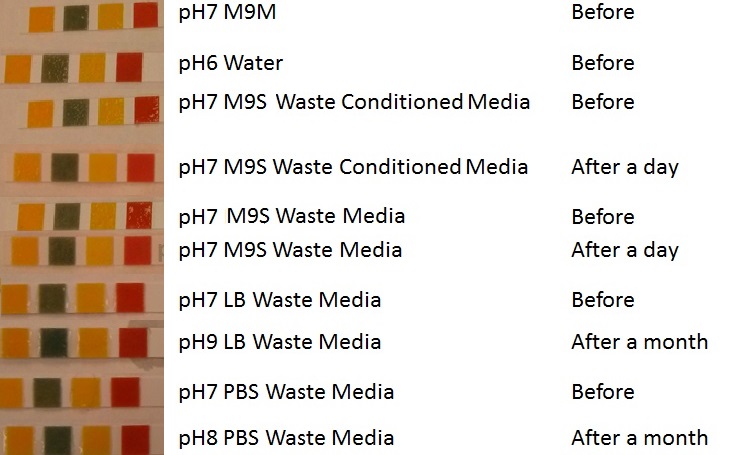
| - | |[[File:Media8888.jpg|thumbnail|right| | + | |[[File:Media8888.jpg|thumbnail|right|450px|<b>pH of experimental media.</b> pH measurements of experimental media were made both before and after several experiments. The time periods refer to the duration MG1655 transformed E. coli were cultured in the media before a pH measurement was made. Figure made by Imperial College London 2013 iGEM.]] |
|} | |} | ||
| - | <b>Conclusion: MG1655 E. coli are viable and grow in all of our experimental medias. We have established a novel media that is optimised for characterisation of biobricks within a mixed waste/landfill context.</b> | + | <b>Conclusion: MG1655 E. coli are viable and grow in all of our experimental medias. We have established a novel media that is optimised for characterisation of biobricks within a mixed waste/landfill context. LB as a long term media suffers from increased alkalinity over a period of a month</b> |
<br> | <br> | ||
| Line 22: | Line 22: | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
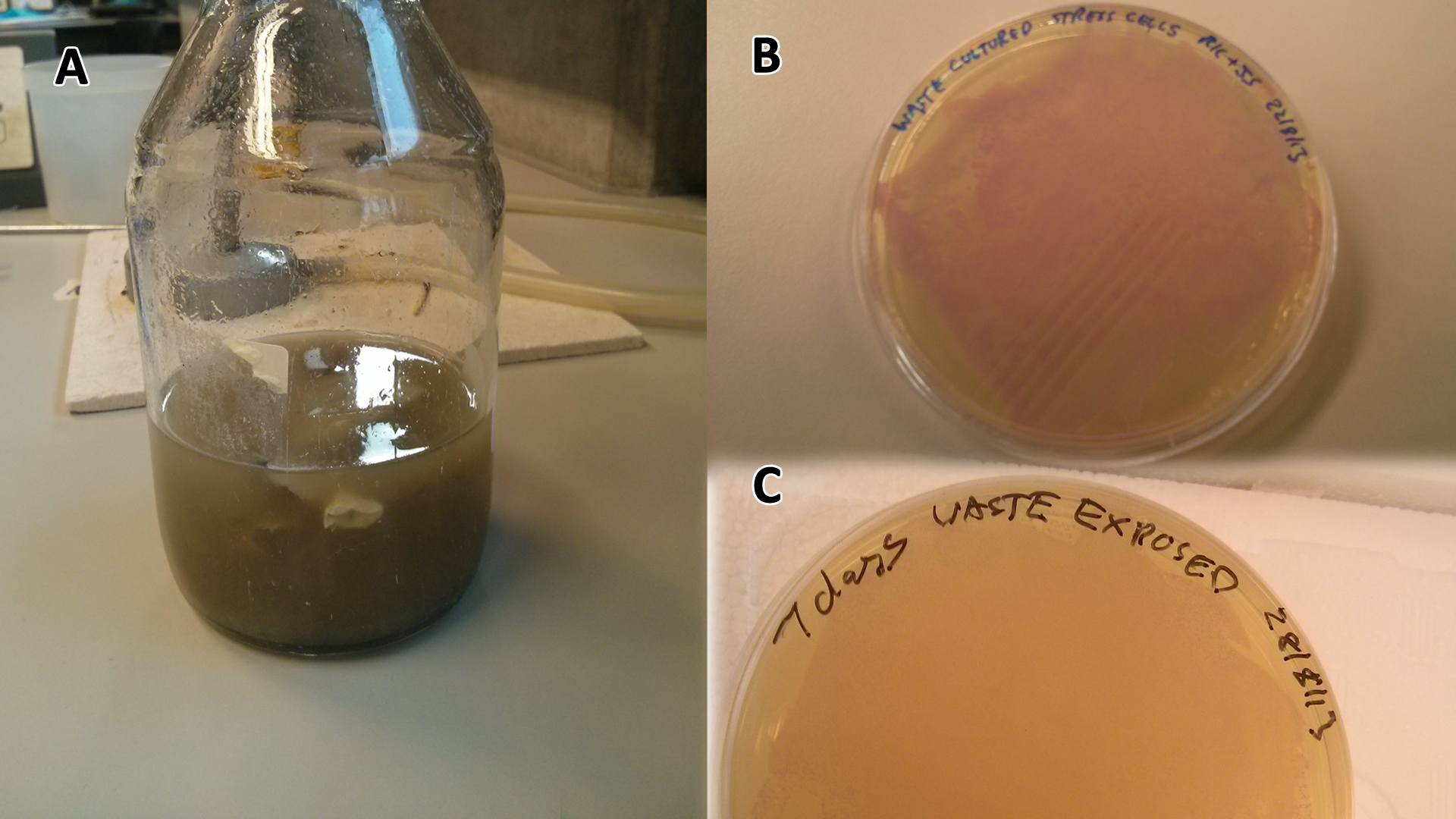
| - | |[[File:Waste_cocktail.png|thumbnail|center| | + | |[[File:Waste_cocktail.png|thumbnail|center|450px|<b>LB-waste assay(A)</b> Waste media <b>(B)</b> Ecoli containing mCherry stress biosensor (BBa_K639003) were grown in mixed waste <b>(A)</b> over 3 days, then streaked in a qualitative assay to check for growth. <b>(C)</b> mCherry stress biosensor (BBa_K639003) transformed Ecoli were streaked again after 7 days growth in SRF. Figure made by Imperial College London 2013 iGEM.]] |
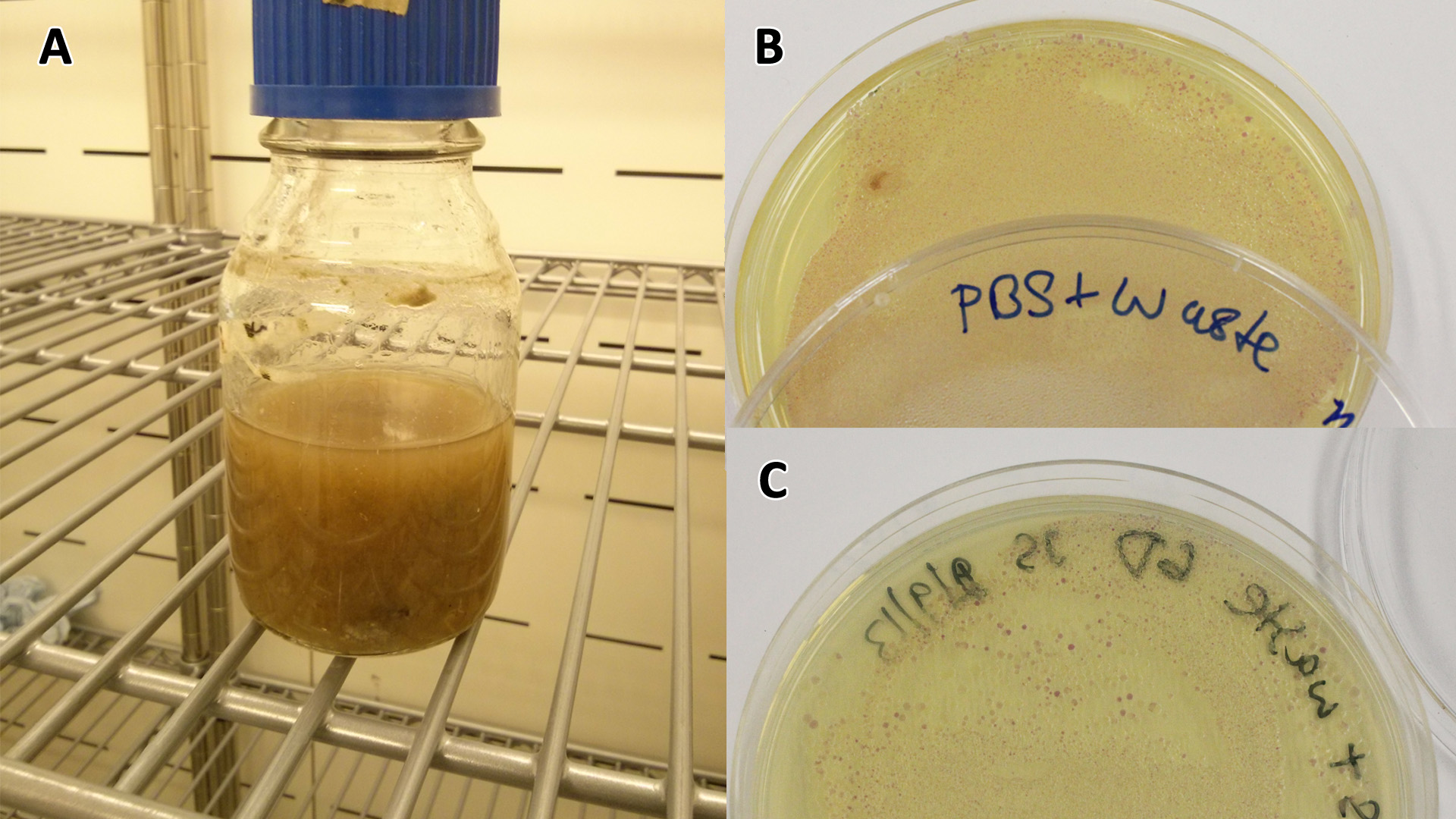
| - | |[[File:PBS_Plus_Waste.jpg|thumbnail|center| | + | |[[File:PBS_Plus_Waste.jpg|thumbnail|center|450px|<b>PBS-waste assay(A)</b> waste media made up in PBS (phosphate buffered saline). <b>(B)</b> E coli expressing mCherry stress biosensor (BBa_K639003) grown in waste media <b>(A)</b> over 3 days, then streaked onto an antibiotic containing plate to qualitatively assess whether the E. coli had survived. <b>(C)</b> Streaked again after 6 days growth in SRF. Figure made by Imperial College London 2013 iGEM.]] |
|} | |} | ||
| Line 34: | Line 34: | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| - | |[[File: | + | |- |
| + | | colspan="2" | | ||
| + | [[File:Very_stressed_ecoli.jpg|thumbnail|left|900px|<b>Photograph of waste conditioned media cultures</b> mCherry stress biosensor (BBa_K639003) transformed MG1655 were grown with [https://2013.igem.org/Team:Imperial_College/Protocols LB-WCM] at 37ºC, with shaking for 48 hours. Figure made by Imperial College London 2013 iGEM.]] | ||
| - | |[[File: | + | |- |
| - | + | |[[File:WCM_media.png|thumbnail|center|450px|<b>Growth assay in waste conditioned media (WCM)</b>. E. coli (MG1655) transformed with [http://parts.igem.org/Part:BBa_K639003 mCherry stress biosensor.] were grown with LB-based waste conditioned media at 37ºC, with shaking. Error bars represent S.E.M., n=4. Figure made by Imperial College London 2013 iGEM.]] | |
| - | [[File:WCM2.png|thumbnail|center| | + | || |
| + | [[File:WCM2.png|thumbnail|center|450px|<b>Growth assay in waste conditioned media (WCM).</b> E. coli (MG1655) transformed with either empty vector control (EV) or mCherry stress biosensor (BBa_K639003) were grown in WCM for 5 hours, with shaking at 37ºC. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
|} | |} | ||
| Line 47: | Line 50: | ||
| - | <h2 class="clear">Growth and induction assays of our Biobricks</h2> | + | <h2 class="clear">Growth and induction assays of our Biobricks in LB</h2> |
| - | Growth and induction assays of our project biobricks. Several of our constructs contain sfGFP within an operon and therefore fluorescence can be utilised to determine if expression is being induced by either addition of Arabinose or Xylose as appropriate to the construct. | + | Growth and induction assays of our project biobricks. Several of our constructs contain sfGFP within an operon and therefore fluorescence can be utilised to determine if expression is being induced by either addition of Arabinose or Xylose as appropriate to the construct. <span style="color:red">'''All of these characterisation assays were undertaken in LB media.'''</span> |
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
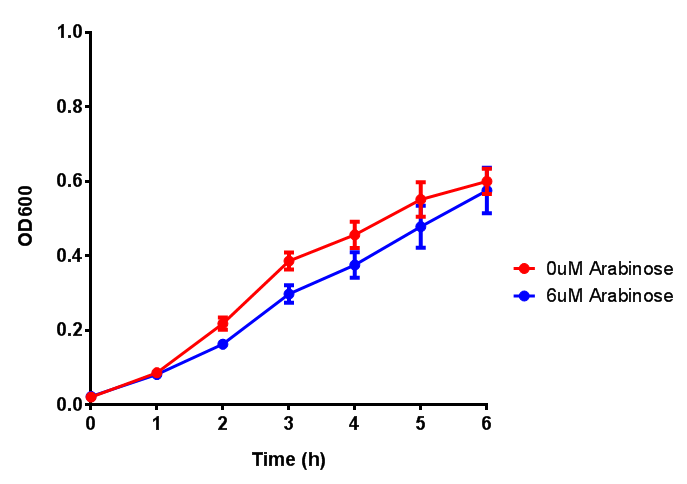
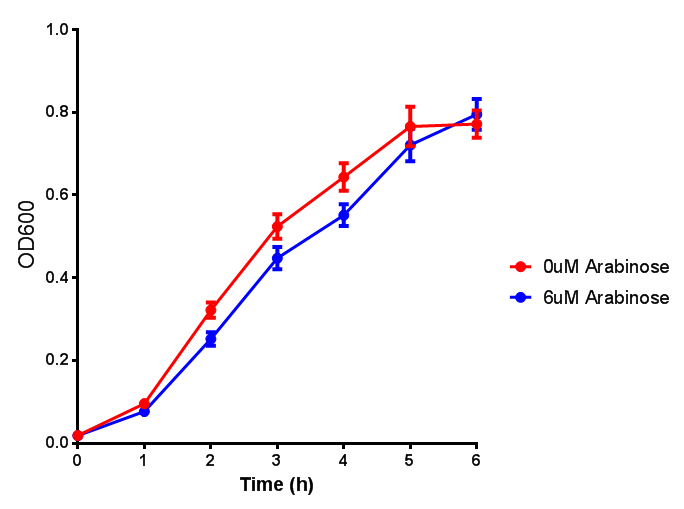
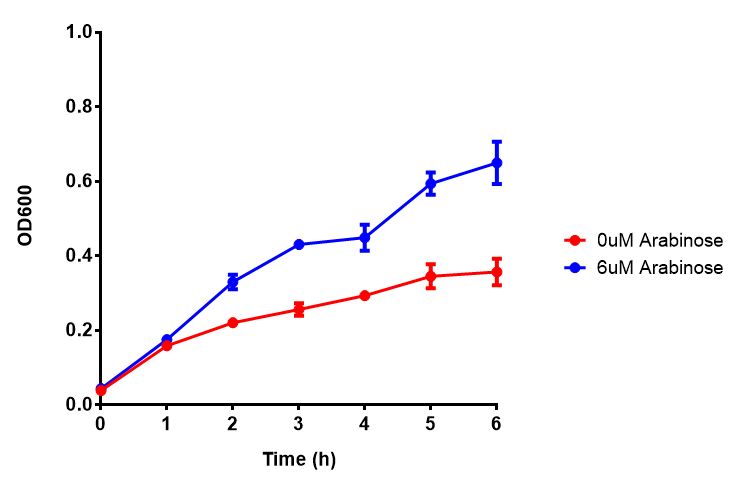
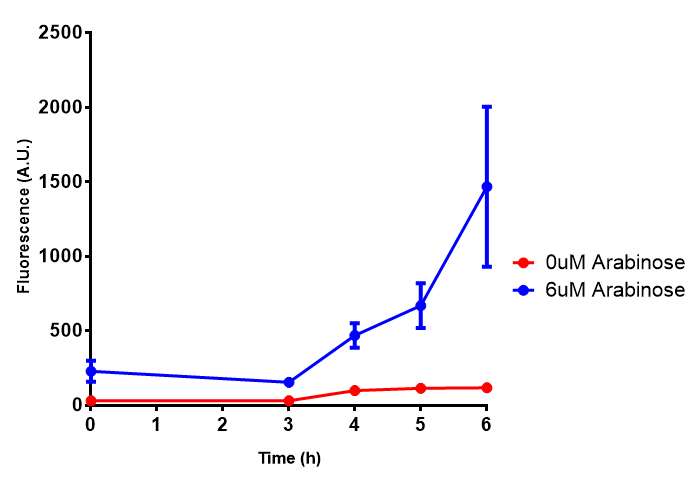
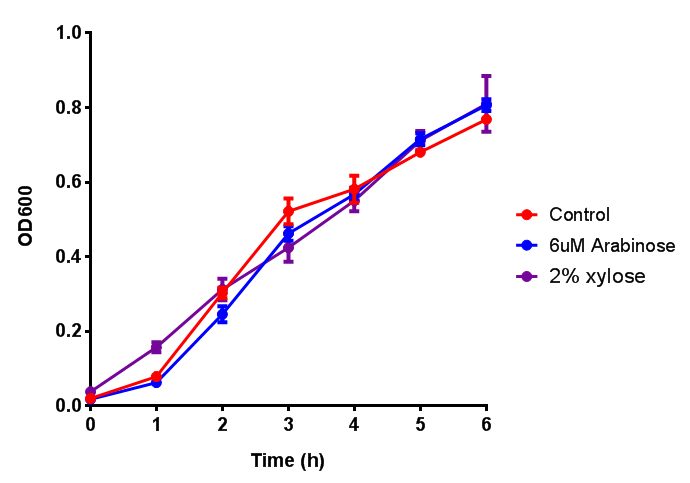
| - | | [[File:Bdh2.png|thumbnail|right| | + | | [[File:Bdh2.png|thumbnail|right|450px|<b>Bdh2 with pelB secretion tag growth assay.</b> ''E. coli'' (MG1655) transformed with pelB-bdh2 [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149013 BBa_K1149013] were grown with either 0 μM or 6 μM Arabinose to induce bdh2 and sfGFP expression. Bdh2 induction shows reduced growth of MG1655 but after 6h they reach the same OD, the decrease in growth is confirmed as not significant by a two-tailed t-test where p = 0.6964 > 0.05 (critical value). Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
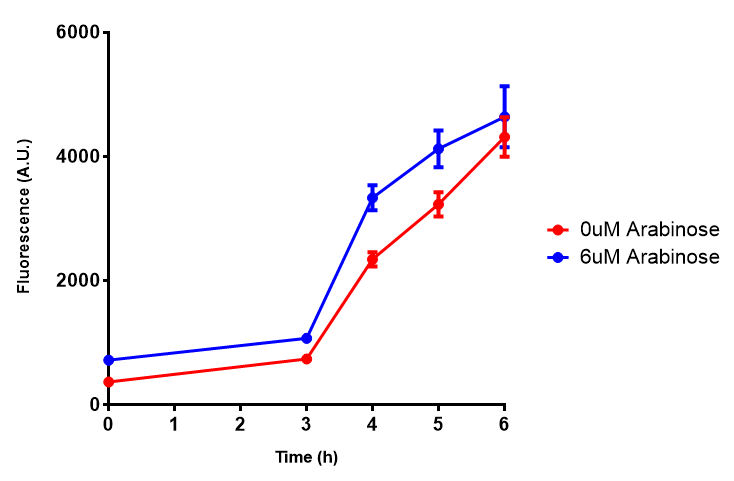
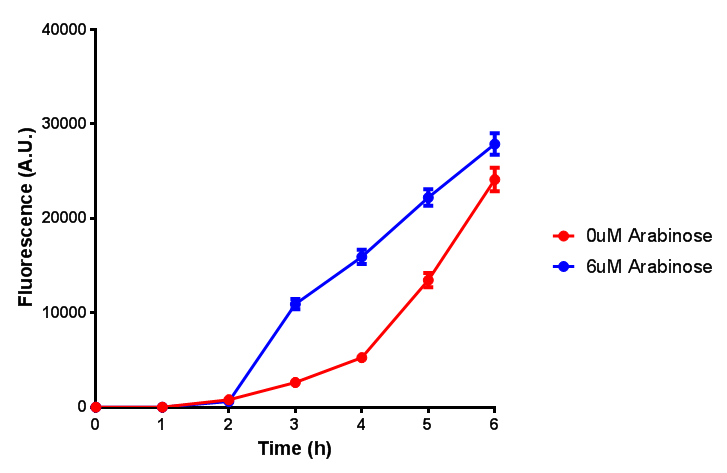
| - | | [[File:Bdh2_fluor.png|thumbnail|right| | + | | [[File:Bdh2_fluor.png|thumbnail|right|450px|<b>Bdh2 with pelB secretion tag induction assay.</b> ''E. coli'' (MG1655) transformed with pelB-bdh2 [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149013 BBa_K1149013] were grown with either 0 μM or 6 μM Arabinose to induce bdh2 and sfGFP expression. We see that induction leads to stronger expression of bdh2 than without, although the promoter is leaky as the curve follows a similar pathway. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
|- | |- | ||
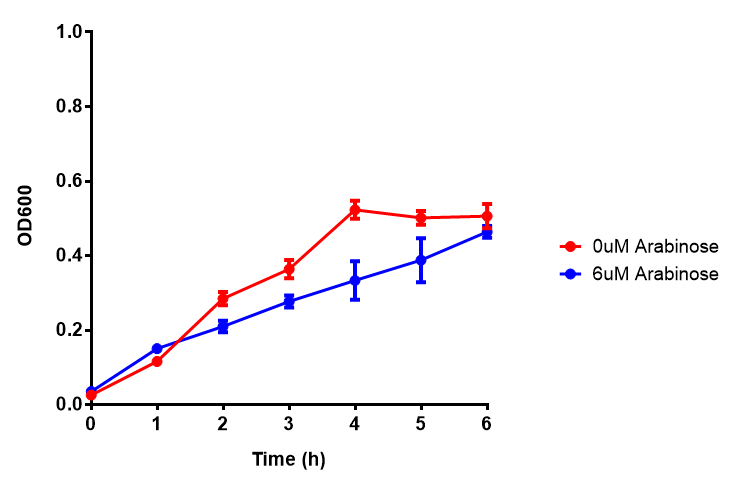
| - | |[[File:Bdh2-2.png|thumbnail|left| | + | |[[File:Bdh2-2.png|thumbnail|left|450px|<b>Bdh2 with no pelB secretion tag growth assay.</b> MG1655 with intracellular bdh2 were grown over 6h to gauge the effect on growth when induced. Graph shows that while they reach the same end point, growth is initially faster without induction for [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149050 BBa_K1149050], though this was not significant as a two-tailed t-test gave p = 0.4930 > 0.05, allowing us to accept the null hypothesis. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| - | |[[File:Bdh2-2_fluor.png|thumbnail|left| | + | |[[File:Bdh2-2_fluor.png|thumbnail|left|450px|<b>Bdh2 with no pelB secretion tag induction assay.</b> In addition to testing [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149050 BBa_K1149050] growth, we looked at induction whereby we see that more sfGFP is produced when induced. Although the promoter is leaky as the curve of intracellular bdh2, as seen with bdh2-pelB is followed tightly by the non-induced cells. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
|- | |- | ||
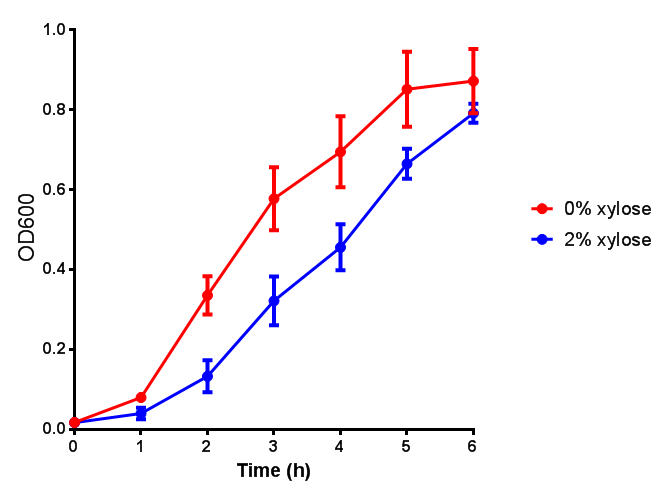
| - | | [[File:CLE.png|thumbnail|right| | + | |
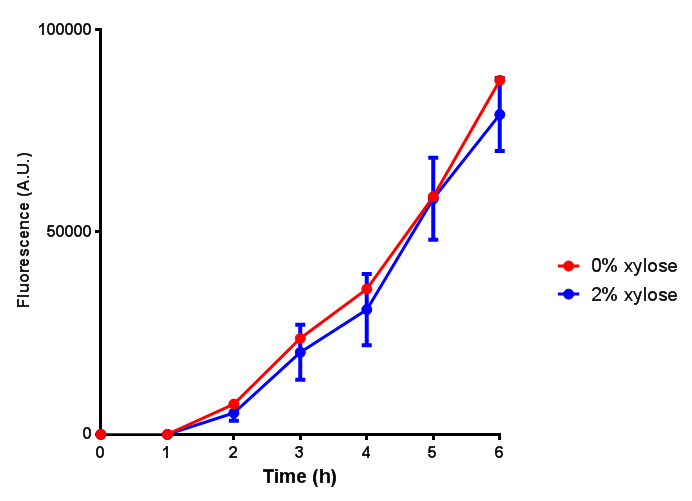
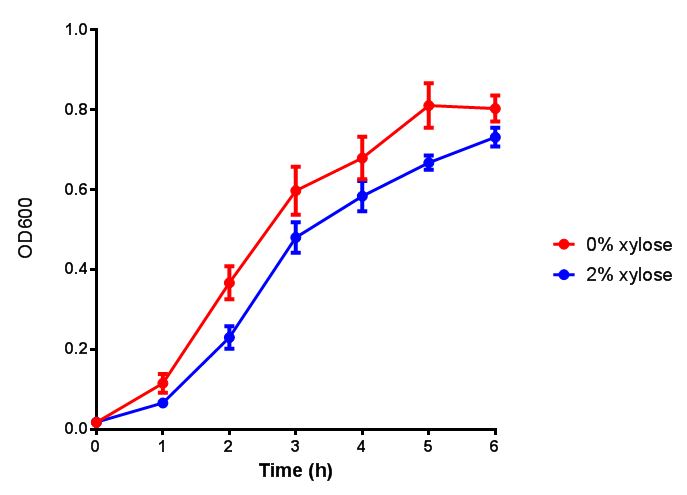
| - | | [[File:CLE_fluor.png|thumbnail|right| | + | | [[File:CLE.png|thumbnail|right|450px|<b>CLE growth assay.</b> <i>E. coli</i> (MG1655) transformed with CLE [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149009 BBa_K1149009] were grown with either 0% or 2% Xylose to induce CLE and sfGFP expression. Induction with xylose shows growth inhibition. Ideally you would normalise the fluorescence with OD600 to give fluorescence/cell in order to get the actual difference in induction expression. A two-tailed t-test shows that p = 0.4308 > 0.05, therefore there is no statistical difference between induced and uninduced. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| + | | [[File:CLE_fluor.png|thumbnail|right|450px|<b>CLE induction assay.</b> <i>E. coli</i> (MG1655) transformed with CLE [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149009 BBa_K1149009] were grown with either 0% or 2% Xylose to induce CLE and sfGFP expression. CLE induction does not seem to increase the amount of fluorescence emitted by the GFP. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |||
|- | |- | ||
| - | | [[File:ESTCS2.png|thumbnail|right| | + | |
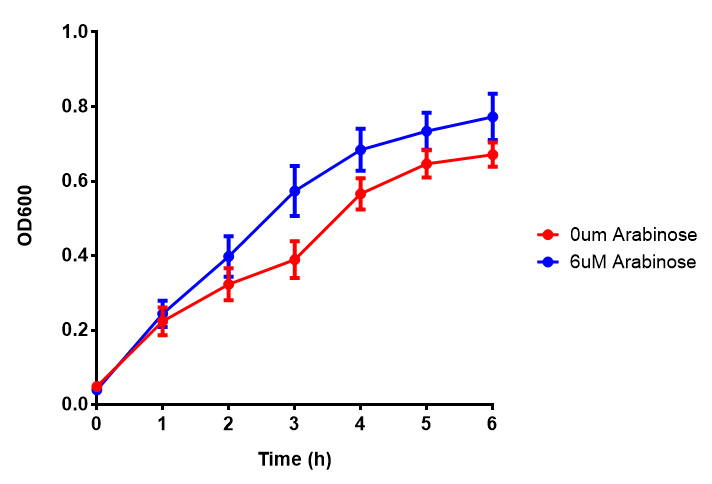
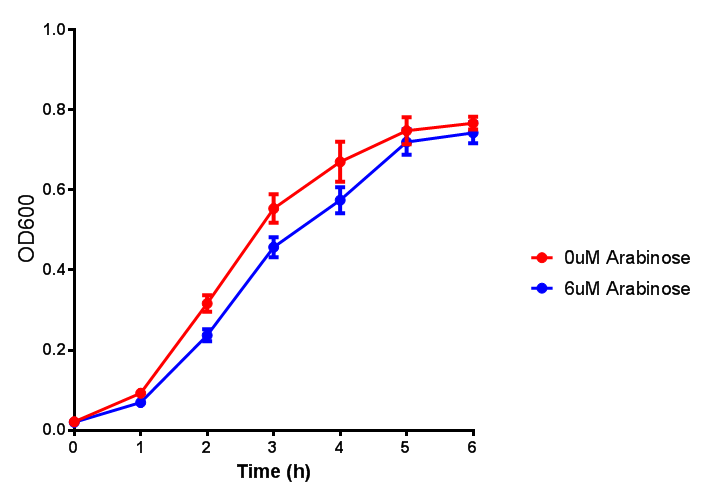
| - | | [[File:ESTCS2_fluor.png|thumbnail|right| | + | |[[File:ESTCS2.png|thumbnail|right|450px|<b>ETSCS2 growth assay.</b> <i>E. coli</i> (MG1655) transformed with ESTCS2 [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149002 BBa_K1149002] were grown with either 0 μM or 6 μM Arabinose to induce ESTC2 and sfGFP expression. The growth curves appear very similar, which is confirmed by a two-tailed t-test p-value = 0.8118, thus no reason to say that there is any growth inhibition. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| + | |[[File:ESTCS2_fluor.png|thumbnail|right|450px|<b>ETSCS2 induction assay.</b> <i>E. coli</i> (MG1655) transformed with ESTCS2 [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149002 BBa_K1149002] were grown with either 0uM or 6uM Arabinose to induce ESTC2 and sfGFP expression. Induction by Arabinose produces a strong response, with induced transformants fluorescing more. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
|- | |- | ||
| - | |[[File:PueA.png|thumbnail|right| | + | |[[File:PueA.png|thumbnail|right|450px|<b>PueA growth assay.</b> MG1655 <i>E. coli</i> were grown for 6h with 0 μM or 6 μM Arabinose. There was no difference between no induction and induction as a t-test gave p = 0.5559 > 0.05. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
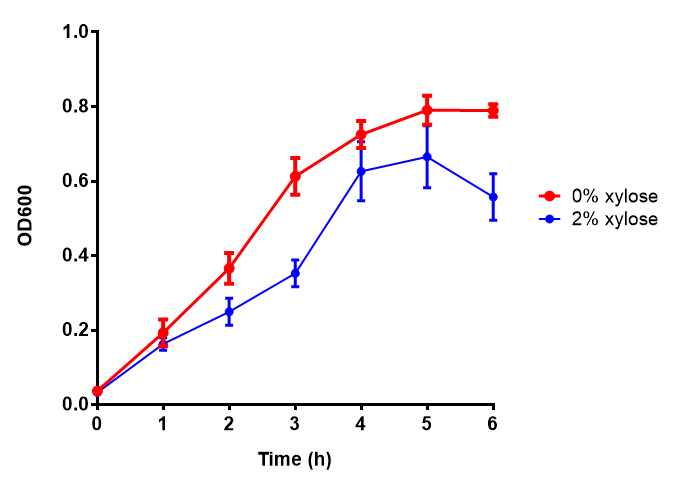
| - | |[[File:PueB.png|thumbnail|right| | + | |[[File:PueB.png|thumbnail|right|450px|<b>PueB growth assay.</b> <i>E. coli</i> (MG1655) transformed with PueB [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149004 BBa_K1149004] were grown with either 0% or 2% Xylose to induce PueB expression. There was no difference between no induction and induction as a t-test gave p = 0.6040 > 0.05. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
|- | |- | ||
| - | |[[File:phaz1.png|thumbnail|left| | + | |[[File:phaz1.png|thumbnail|left|450px|<b>Phaz1 growth assay.</b> MG1655 <i>E. coli</i> were grown for 6h with 0% and 2% Xylose to induce Phaz1 [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149010 BBa_K1149010]. A two-tailed t-test, with p = 0.4181 shows that there is no significant difference between induction and no induction with regards to growth inhibition, as the value exceeds the critical value of 0.05. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| - | |[[File:PulA.png|thumbnail|left| | + | |[[File:PulA.png|thumbnail|left|450px|<b>PulA growth assay.</b> <i>E. coli</i> (MG1655) transformed with PulA (BBa_K1149006) were grown with either 0 μM or 6 μM Arabinose to induce PulA expression. There was no growth inhibition as p = 0.7648 > 0.05, therefore induction did not cause significant growth decrease. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
|- | |- | ||
| - | |[[File:PK.png|thumbnail|left| | + | |[[File:PK.png|thumbnail|left|450px|<b>Proteinase K growth assay.</b> MG1655 <i>E. coli </i> display strong growth with the induction of [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149007 BBa_K1149007], though it is not significant as a two-tailed t-test gave p = 0.1471. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| - | + | |[[File:PK_fluor.png|thumbnail|left|450px|<b>Proteinase K induction assay.</b> When induced, there is a very clear increase in fluorescence for [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149007 BBa_K1149007], however t-test analysis gives p = 0.0595 > 0.05 for this, thus the fluorescence does not significantly differ from uninduced. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | |
| - | + | ||
| - | + | ||
| - | |[[File: | + | |
| - | + | ||
|} | |} | ||
| + | <!-- | ||
| + | |[[File:PK_pelB.png|thumbnail|left|450px|<b>Proteinase K with pelB secretion tag growth assay.</b> As with Proteinase K without pelB, there is increased growth compared to uninduced for [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149008 BBa_K1149008]. However this is not significant as a two-tailed t-test gave p = 0.4442. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |[[File:PK_pelB_fluor.png|thumbnail|left|450px|<b>Proteinase K with pelB secretion tag induction assay.</b> Induction of [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149008 BBa_K1149008] gives a very strong induction response, this is significant as a two-tailed t-test gave a value of p = 0.0104 < 0.05. Thus the induction does have a significant effect on fluorescence for Proteinase K with pelB. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | --> | ||
| - | <h3 | + | <h3 class="clear">Empty Vector Control</h3> |
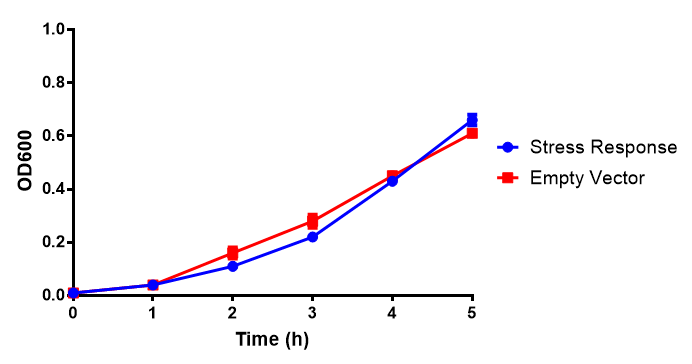
| - | [[File:EV2.png|thumbnail|600px|<b>Empty vector (EV) induction assay</b> E. coli (MG1655) transformed with empty vector (EV) control plasmid were grown with either 0uM or 6uM Arabinose. Growth was at | + | [[File:EV2.png|thumbnail|center|600px|<b>Empty vector (EV) induction assay</b> E. coli (MG1655) transformed with empty vector (EV) control plasmid were grown with either 0uM or 6uM Arabinose. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| + | <b>Conclusions: Extracellular proteinase K fluoresces significantly more than uninduced extracellular proteinase K. None of the biobricks had significant decreases in their growth rates when induced by the respective inducer - be that Arabinose or Xylose. The remainder of the fluorescence assays did not show a significant difference between induction and no induction. This said, the data still requires normalising, this process might elucidate finally as to whether any other biobricks have significant fluorescence when induced. The empty vector control whether induced or not shows similar growth, proving its functionality as a control.</b> | ||
| + | <br> | ||
| + | <h1 class="clear">Growth assay characterisation of existing biobricks</h1> | ||
| - | <h2 | + | <h2>Stress biosensor characterisation (BBa_K639003)</h2> |
| - | + | ||
| - | + | ||
Originally we intended on using [http://parts.igem.org/Part:BBa_K639003 BBa_K639003] to detect whether our cells were stressed when grown with an array of potentially toxic plastics and degradation products. However, as the data below shows, the promoter is leaky and expresses mCherry in a non stressed state. As an alternative we utilised the stress sensor as a marker for our chassis E. coli (MG1655) for an array of qualitative and quantitative waste growth and toxicity assays. | Originally we intended on using [http://parts.igem.org/Part:BBa_K639003 BBa_K639003] to detect whether our cells were stressed when grown with an array of potentially toxic plastics and degradation products. However, as the data below shows, the promoter is leaky and expresses mCherry in a non stressed state. As an alternative we utilised the stress sensor as a marker for our chassis E. coli (MG1655) for an array of qualitative and quantitative waste growth and toxicity assays. | ||
| Line 93: | Line 100: | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
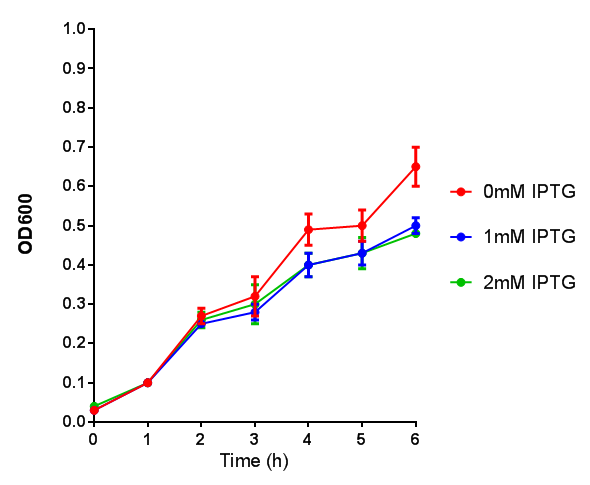
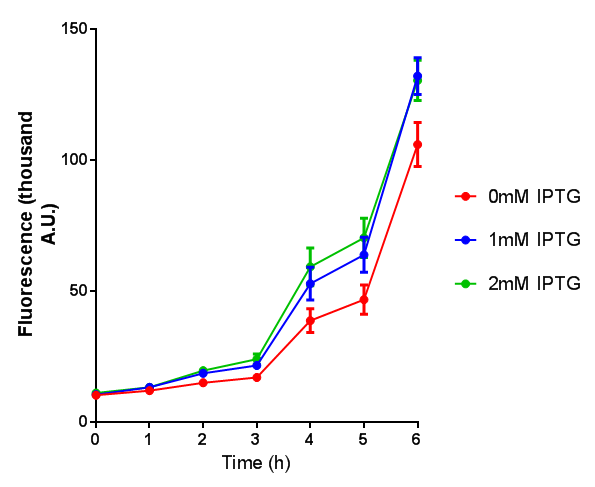
| - | |[[File:OD_stress.png|thumbnail|right| | + | |[[File:OD_stress.png|thumbnail|right|450px|<b>Stress sensor growth assay.</b> <i>E. coli</i> (MG1655) transformed with stress biosensor [http://parts.igem.org/Part:BBa_K639003 BBa_K639003]. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| - | |[[File:Fluorescence_stress.png|thumbnail|right| | + | |[[File:Fluorescence_stress.png|thumbnail|right|450px|<b>Stress sensor IPTG induced fluorescence.</b> <i>E. coli</i>(MG1655) transformed with stress biosensor [http://parts.igem.org/Part:BBa_K639003 BBa_K639003]. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
|- | |- | ||
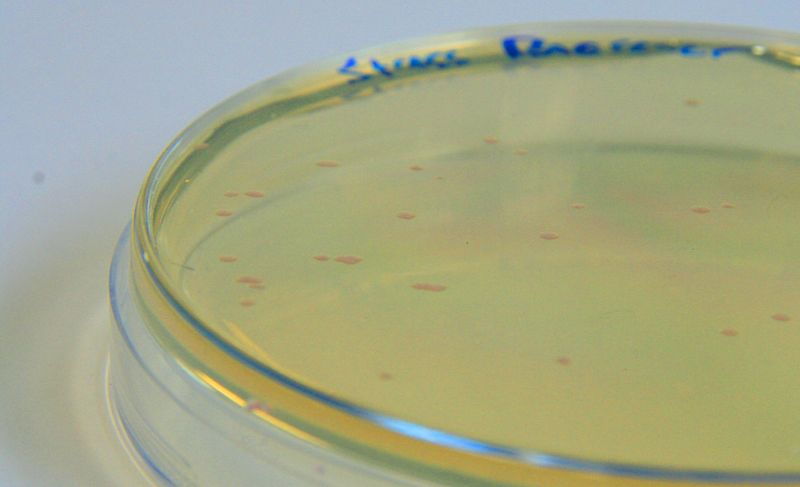
| - | |[[File:StressResponse.jpg| | + | |[[File:StressResponse.jpg|450px|thumb|left|<b>[http://parts.igem.org/Part:BBa_K639003 BBa_K639003] transformed into <i>E. coli</i>. strain MG1655.</b> Pink colonies are visible, which relate to 'leaky' mCherry production]] |
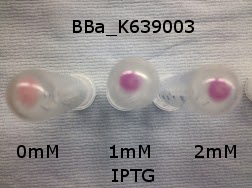
| - | |[[File:K639003IPTG.jpeg| | + | |[[File:K639003IPTG.jpeg|thumbnail|center|450px|<b>[http://parts.igem.org/Part:BBa_K639003 BBa_K639003] transformed into <i>E. coli</i>. strain MG1655.</b> Cells were grown at 37°C in 4ml LB with 0, 1 or 2mM IPTG. At 6 hours post IPTG induction, cells were spun down and imaged.]] |
|} | |} | ||
| + | <b>Conclusion: The stress biosensor is activated when induced with IPTG but is leaky. In addition, growth decreases when cells are induced</b> | ||
| + | <br> | ||
| + | <h2>phaCAB biobrick characterisation</h2> | ||
| + | <h3 class="clear">LB</h3> | ||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
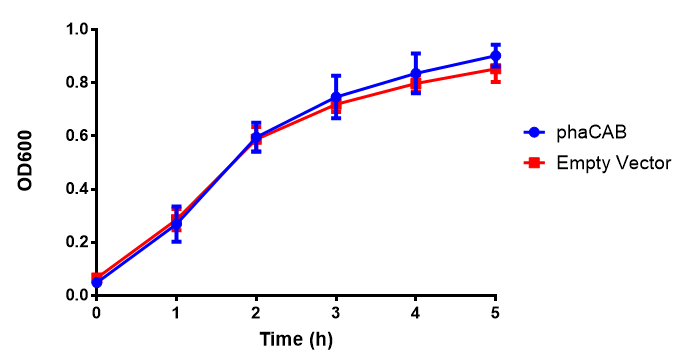
| + | |[[File:LB.png|thumbnail|center|450px|<b>MG1655 in LB with plasmids EV and phaCAB ([http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149041 BBa_K1149041]).</b> There is no growth inhibition when comparing the empty vector with the phaCAB vector in LB media. LB shows the strongest growth curve with minimal lag phase. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
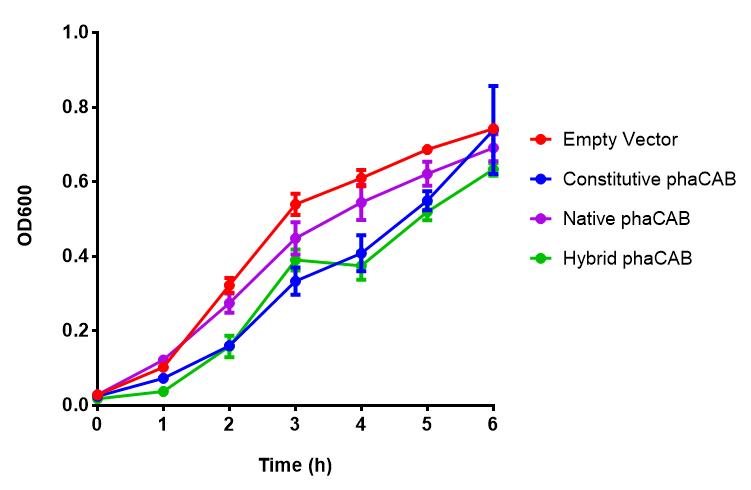
| - | < | + | |[[File:PhaCAB_LB.png|thumbnail|center|450px|<b>MG1655 in LB with plasmid EV and the native, constitutive and hybrid promoters for phaCAB.</b> The plasmids all show similar growth curves in LB for all 3 promoters. Indeed this is confirmed by an ANOVA test, which shows no significance in terms of growth difference the constructs (<i>F</i> <sub>3,24</sub> = 0.3573, p < 0.7843). Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| + | |} | ||
| - | < | + | <h3 class="clear">M9 Minimal</h3> |
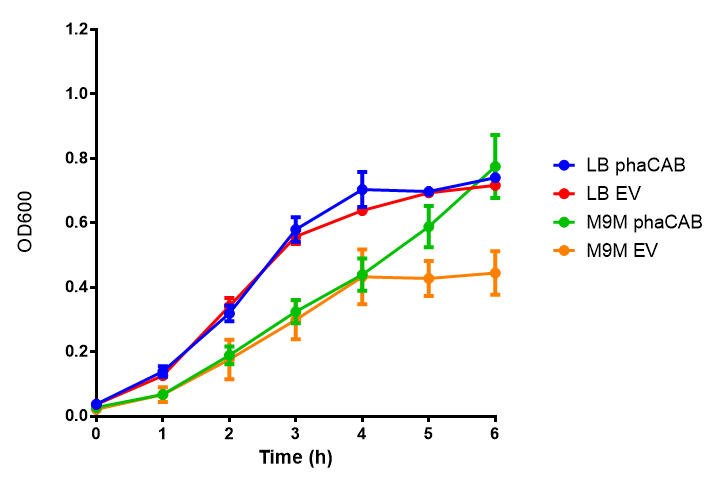
| - | [[File: | + | [[File:M9M.png|thumbnail|center|500px|<b>MG1655 in M9M with plasmids EV and phaCAB.</b> There is no growth inhibition when comparing the empty vector with the phaCAB vector in minimal media. M9M shows the least growth growth of all the media as it has low carbon and amino acid content. Growth was at 37°C with shaking over 5h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| - | < | + | <h3 class="clear">M9 Supplemented</h3> |
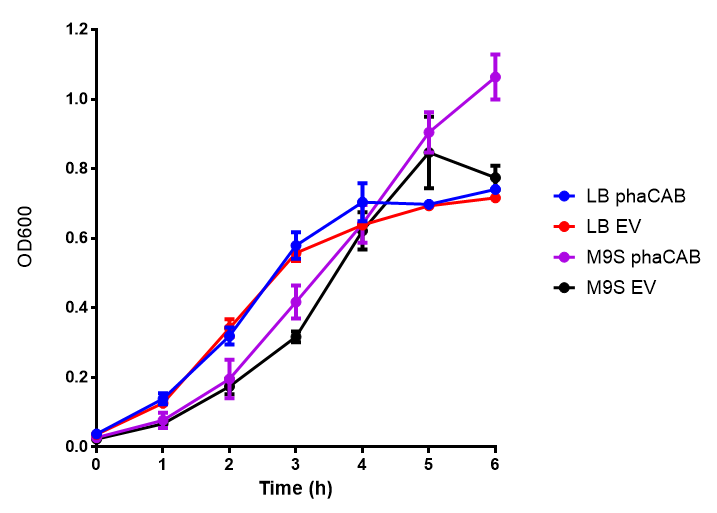
| - | [[File: | + | [[File:M9S.png|thumbnail|center|500px|<b>MG1655 in M9S with plasmids EV and phaCAB.</b> There is no growth inhibition when comparing the empty vector with the phaCAB vector in supplemented minimal media. M9S shows a lag phase in growth but quickly increases due to increased amino acid content, nearly reaching LB after 5h. Growth was at 37°C with shaking. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| - | + | <b>Conclusions: No difference between any of our phaCAB promoter constructs (native, hybrid, constitutive) when grown on LB compared to empty vector. In addition, there was no marked difference for native phaCAB in M9 minimal and M9 supplemented media, although the <i>E. coli</i> grew very well in supplemented idea.</b> | |
| - | + | <br><br> | |
| + | <h2 class="clear">pBAD characterisation</h2> | ||
| + | Characterisation of the strong pBAD promoter was a necessity to our project. We wanted to know whether or not it was inducing a significant increase in fluorescence of our constructs. We also wanted to know whether arabinose at the concentration we were inducing with (6 μM) was sufficient. Finally we wanted to look at whether or not glucose has an effect on growth. In order to address this we performed assays with pBAD in intracellular bdh2 in M9 minimal media. To tie it in with our project, we also used M9 minimal waste conditioned media (M9MWCM) to compare growth. Bdh2 is responsible for the synthesis of acetoacetate from 3HB and tagged with sfGFP. | ||
| - | < | + | <!-- |
| + | |||
| + | The null hypothesis, arabinose concentration does not influence growth of MG1655 in M9 minimal (M9M) media and M9 minimal waste conditioned media (M9MWCM) was tested using a two-tailed t-test. This must be rejected because p < 0.0217, thus arabinose concentration has an influence on growth in M9M and M9MWCM. | ||
| + | --> | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
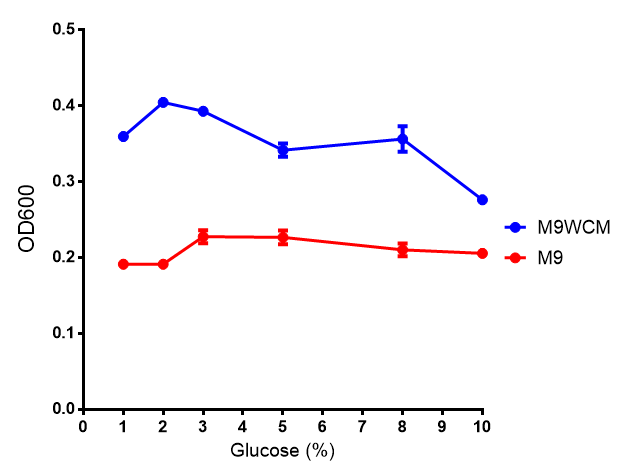
| - | | [[File: | + | | [[File:Glucose_inhibition.png|thumbnail|right|450px|<b>Glucose inhibition on growth in minimal media on bdh2.</b> The outcome of this growth assay shows that cells grow more in M9WCM but that increasing glucose concentration influences growth in M9WCM while it is unaffected in M9 minimal media. Characterisation was done in M9 and M9WCM. Data points show final time point after 6h growth for each concentration. The arabinose concentration was kept at 6 μM for all samples. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| - | | [[File: | + | |
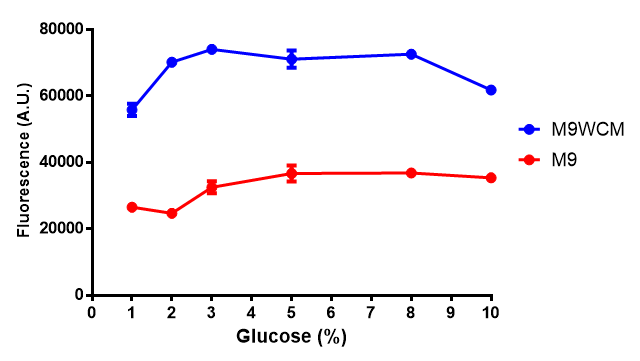
| + | | [[File:F_glucose_inhibition.png|thumbnail|right|450px|<b>Glucose inhibition on fluorescence in minimal media on bdh2.</b> The fluorescence of GFP in bdh2 under M9WCM shows significant difference in fluorescence output (p < 0.0001.). This compared to M9 media where fluorescence is almost half. The fluorescence in glucose is quite stable despite the decreased growth witnessed in the growth assay. Characterisation was done in M9 and M9WCM. Data points show final time point after 6h growth for each concentration. The arabinose concentration was kept at 6 μM for all samples. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
|} | |} | ||
| + | The null hypothesis, glucose concentration does not influence growth of MG1655 in M9 minimal (M9M) media and M9 minimal waste conditioned media (M9MWCM) was tested using a two-tailed t-test. This must be rejected because p < 0.0001, thus glucose concentration has an influence on growth in M9M and M9MWCM. In addition to this, the null hypothesis: glucose concentration does not influence fluorescence in M9 minimal media must be rejected as p < 0.0001. Thus glucose concentration influences fluorescence. | ||
| + | |||
| + | <br><br> | ||
| + | <b>Conclusions: Growth is significantly influenced glucose with regards to growth in M9M and M9MWCM. <!--<span style="color:#f00">Bdh2 grown in waste have an optimum at 10 μM arabinose</span> while with 2 μM arabinose when grown in M9M, they are already at optimum.--> <span style="color:#f00">Bdh2 grown in glucose grow better in M9MWCM than M9M but are inhibited with increasing glucose concentration.</span></b> | ||
| + | <br> | ||
| + | |||
| + | <h2>Glucose </h2> | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
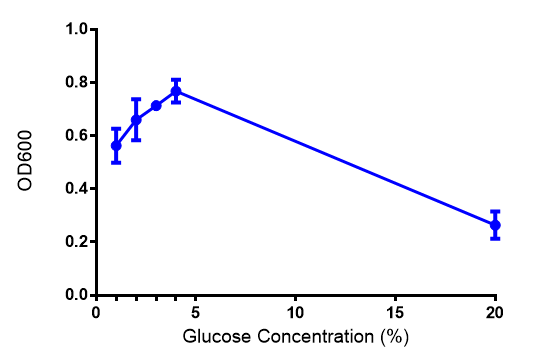
| - | | [[File: | + | |[[File:Glucose_graph.png|thumbnail|center|450px|<b>Effect of glucose concentration on growth of phaCAB.</b> Cell growth of phaCAB in<i>E. coli</i> at 5 glucose concentrations 1%,2%,3%,4% and, 20% (55 mM, 111 mM, 167 mM, 222 mM and 1.1M respectively). Optimum growth is at 2-4% glucose. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| - | | [[File: | + | |
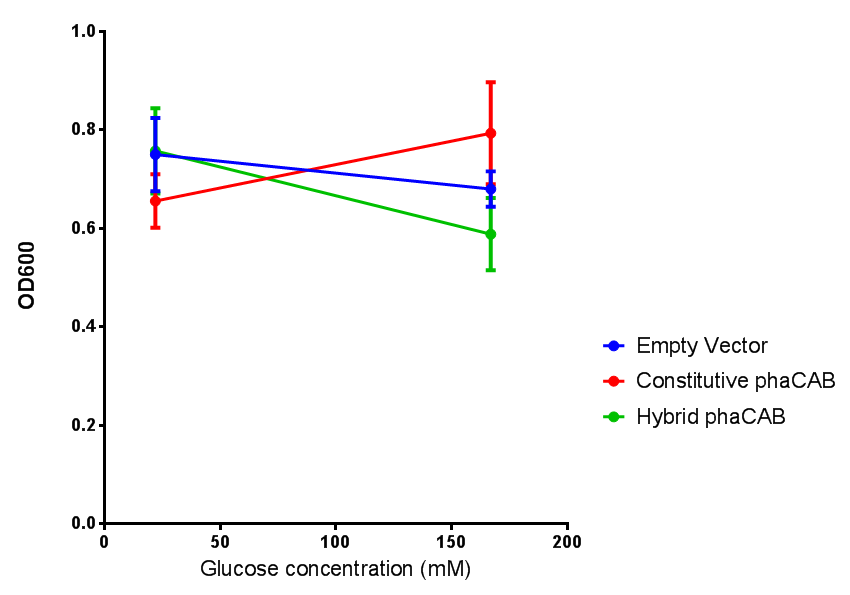
| + | |[[File:(M9S_glucose).png|thumbnail|center|450px|<b> phaCAB growth in M9S supplemented with glucose.</b> Constitutive phaCAB grows more at higher glucose concentration of 167 mM while the hybrid grows better at a lower glucose concentration of 22 mM. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |- | ||
| + | | [[File:3%25_glu_M9M.png|thumbnail|right|450px|<b> phaCAB growth in M9 minimal with 3% glucose sole carbon source.</b> LB grown MG1655 phaCAB grow more rapidly initially then M9M with 3% glucose sole carbon source but reach the same OD after 6h while EV shows a different trend. EV in M9M levels off at a much lower OD at 4h, as seen with EV grown in LB. ANOVA analysis shows that the null hypothesis that there is no significant difference between M9M and LB in empty vector and phaCAB, thus we must accept the null hypothesis (<i>F</i> <sub>3,24</sub> = 0.8451, p < 0.4827). Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |||
| + | | [[File:3%25_glu_M9S.png|thumbnail|right|450px|<b> phaCAB growth in M9S supplemented with 3% glucose.</b> LB grown MG1655 phaCAB grow more rapidly initially then M9S with 3% glucose sole carbon source, but after 5h, phaCAB in M9S continue to grow to a higher OD. EV shows a different trend, in M9S it levels off at a similar level to LB. ANOVA analysis for the null hypothesis that there is no significant difference between M9S and LB similarly must be rejected as p > 0.05 (<i>F</i> <sub>3,24</sub> = 0.9802, p < 0.06009). Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
|} | |} | ||
| + | <b>Conclusions: Glucose has an optimum growth around 3-4%. In M9S, it does not give any growth advantage to cells cultured in higher glucose concentrations. Cells grown in M9S grew equally well as those grown in LB, possibly because of LB having a pH shift as it gets used up.</b> | ||
| - | <h3> | + | |
| - | [[File: | + | <h2>Plastic Toxicity Assays</h2> |
| + | |||
| + | <h3>L-lactic Acid </h3> | ||
| + | [[File:L-LActic_Acid.png|thumbnail|center|600px|<b> L-lactic acid in LB with Stress Response cells.</b> MG1655 cells were grown on 0 mM and 5 mM L-Lactic Acid. Lactic acid is a byproduct of polylactic acid degradation, a key component to our project. Cells grown in 5mM L-lactic acid show a pronounced lag phase before commencing log growth. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | <h3>Ethylene glycol </h3> | ||
| + | |||
| + | [[File:EG_growth.png|thumbnail|center|600px|<b> Ethylene glycol in LB with Stress Response cells.</b> MG1655 were grown in ethylene glycol, a byproduct of polyurethane degradation. Cells were grown in 0mM, 100mM or 200mM Ethylene Glycol. At 37°C, the concentrations of ethylene glycol used do not affect growth, however at 30°C, the increasing concentration results in halved growth. A two-tailed t-test addressing the null hypothesis, temperature does not affect growth with ethylene glycol shows that the null hypothesis must be rejected as p = 0.001. Data points show final time point after 6h growth for each concentration. Growth was at 37°C and 30°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |||
| + | Reduced growth at 30°C likely due to decreased efficiency of MG1655 ethylene glycol break down enzymes. These enzymes (see UC Davis 2012) are endogenously expressed and detoxify Ethylene Glycol. | ||
| + | |||
| + | <h3>3-hydroxybutyrate (3HB) </h3> | ||
| + | [[File:3HB666.png|thumbnail|center|600px|<b> Toxicity of 3-hydroxybutyrate in LB with phaCAB and EV expression.</b> 3HB was tested at 5 concentrations for toxicity - 1 μM, 100 μM, 1 mM, 10 mM and 237 mM. Between 10 mM and 237 mM, there is a clear toxicity effect observed. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | <br><br> | ||
| + | [[File:Imperial_3HBtoxicity_bdh2OD.jpg|thumbnail|center|600px|<b>Growth reduction effect of 3-hydroxybutyrate in LB on bdh2 and EV. </b>3HB was tested at 4 concentrations for toxicity - 1 mM, 10 mM, 50 mM and 100 mM. Between 10 mM and 50 mM, there is a slight growth inhibitory effect observed in bdh2 cells that were induced by 1 mM arabinose. Data points show final time point after 7h growth for each concentration. Growth was at 37°C with shaking over 7h. Error bars are SEM, n=3. Figure made by Imperial College London 2013 iGEM.<br><b><span style="color:#FF8000"><font size="6">Performed after European Jamboree</font size="6"></span></b>]] | ||
| + | |||
| + | |||
| + | <br><br> | ||
| + | [[File:Imperial_3hbtoxicity_bdh.jpg|thumbnail|center|600px|<b>Protein expression of MG1655 with bdh2 in 3-hydroxybutyrate in LB. </b>3HB was tested at 4 concentrations for induced protein expression assay - 1 mM, 10 mM, 50 mM and 100 mM. 3-hydroxybutyrate might have reduced protein expression as indicated by fluorescence/OD600 (t-test p <0.05). Fluorescence sensitivity was set to be 50. Data points show final time point after 7h growth for each concentration. Growth was at 37°C with shaking over 7h. Error bars are SEM, n=3. Figure made by Imperial College London 2013 iGEM.<br><b><span style="color:#FF8000"><font size="6">Performed after European Jamboree</font size="6"></span></b>]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h3>Acetoacetate </h3> | ||
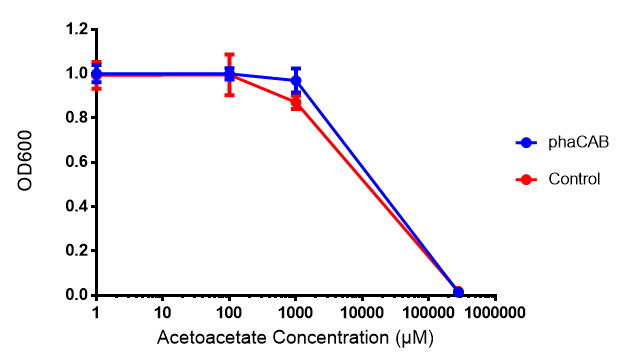
| + | [[File:AA777.png|thumbnail|center|600px|<b> Toxicity of acetoacetate in LB with phaCAB and EV expression.</b> Acetoacetate was also tested for toxicity at 4 concentrations - 1 μM, 100 μM, 1mM and 287 mM. In phaCAB and EV this manifested itself between 1 mM and 287 mM. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |||
| + | <h3>Poly(3-hydroxybutyrate) P(3HB) </h3> | ||
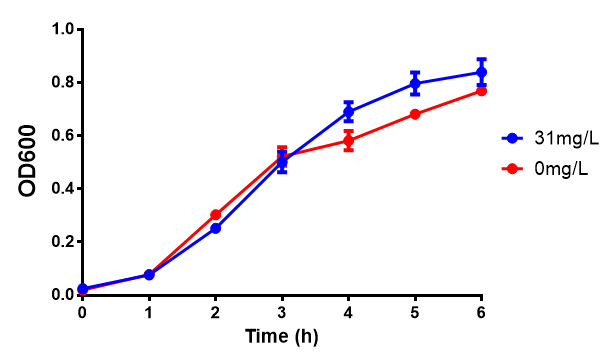
| + | [[File:31ug_P3HB.png|thumbnail|center|600px|<b> Toxicity of Poly(3-hydroxybutyrate) in LB with phaCAB and EV expression.</b> P3HB toxicity was only tested at one concentration in LB - 31 μg/L, it was also tested at 31 mg/L however the resulting OD was over 1, so this data was not considered accurate. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | <br><br> | ||
| + | |||
| + | [[File:Imperial_p3hbtoxicity_od.jpg|thumbnail|center|600px|<b>Growth reduction of Poly(3-hydroxybutyrate) in LB with bdh2 and EV expression.</b>P3HB was tested at 4 concentrations for toxicity - 40 ug/L, 80 ug/L, 400 ug/L and 800 ug/L. These concentrations did not have significant inhibitory effect on cell growth for both bdh2 and empty vector. Bdh2 expression was induced by 1 mM arabinose. Data points show final time point after 7h growth for each concentration. Growth was at 37°C with shaking over 7h. Error bars are SEM, n=3. Figure made by Imperial College London 2013 iGEM.<br><b><span style="color:#FF8000"><font size="6">Performed after European Jamboree</font size="6"></span></b> | ||
| + | ]] | ||
| + | |||
| + | <br><br> | ||
| + | |||
| + | [[File:Imperial_p3hb_bdh2.jpg|thumbnail|center|600px|<b>Protein expression of MG1655 with bdh2 in 3-hydroxybutyrate in LB.</b>P3HB was tested at 4 concentrations for protein expression assay - 40 ug/L, 80 ug/L, 400 ug/L and 800 ug/L. P3HB might have reduced Bdh2 expression at concentration higher than 40 ug/L, as indicated in a t-test comparing the fluorescence/OD600 values (p<0.05). Bdh2 expression was induced by 1 mM arabinose. Fluorescence sensitivity was set at 50. Data points show final time point after 7h growth for each concentration. Growth was at 37°C with shaking over 7h. Error bars are SEM, n=3. Figure made by Imperial College London 2013 iGEM.<br><b><span style="color:#FF8000"><font size="6">Performed after European Jamboree</font size="6"></span></b>]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h3>Poly(lactic acid) (PLA) </h3> | ||
| + | [[File:PLA.png|thumbnail|center|600px|<b> Toxicity of Poly(lactic acid) in LB with phaCAB and EV expression.</b> Polylactic acid in the form of ground up plastic did not show any toxicity effect at 31 mg/L in LB. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |||
| + | <b>Conclusions: Elevated lactic acid results in a pronounced lag phase. At 37°C ethylene glycol does not have a significant effect on MG1655 growth. 3HB and acetoacetate are not toxic until their molarity is or exceeds 237 mM and 287 mM respectively. At the tested concentrations, neither PLA nor P(3HB) had a significant effect on growth. </b> | ||
| + | |||
| + | <h2>Sole carbon source</h2> | ||
| + | |||
| + | <h3>bdh2 improves growth on 3HB</h3> | ||
| + | |||
| + | We have previously observed that MG1655 cells can survive in minimal media that contains 3HB as sole carbon source. This is evidence for that some uptake mechanism and a metabolic pathway is active at a low level in the cells. We hypothesised that the 3HB dehydrogenase could improve growth since it can convert 3HB into acetoacetate, a common metabolite in ''E.coli''. | ||
| + | |||
| + | In order to test this, we run a growth experiment with various 3HB concentrations where the growth of cells containing bdh2 (BBa_K1149050) and Empty vector was recorded on a 96 well plate. We have calculated the growth rates from this data and plot it on the graph below. | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| - | | [[File: | + | |[[File:3HB_sole_carbon_source.png|thumbnail|center|450px|<b> 3HB sole carbon source growth in minimal media with phaCAB and EV.</b> MG1655 phaCAB were grown in supplemented and minimal M9 media. Relative to the empty vector control, there was no significant difference in growth as t-test gave p = 0.8072 > 0.05. However, an ANOVA of the data gave (<i>F</i> <sub>3,8</sub> = 6.589, p < 0.0149), thus the null hypothesis, there is no difference between M9M and M9S must be rejected. Indeed if we look closer, we see that both M9M EV and M9S EV are significantly different from another (p = 0.045), as are M9M phaCAB and M9S phaCAB (p = 0.0284). Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| - | | [[File: | + | |
| + | |[[File:3HB_all.png|thumbnail|center|450px|x|<b> 3HB sole carbon source growth in M9S with P3HB synthesis pathway genes.</b> The 3 phaCAB promoter constructs are represented with native phaCAB, constitutive phaCAB and hybrid phaCAB. In addition bdh2 was tested to see whether its presence increased growth for the cells. This was completed with 3 3HB concentrations - 100 μM, 10 mM and 100 mM. ANOVA analysis shows that there is no significant difference between any of the constructs (<i>F</i> <sub>4,10</sub> = 0.1875, p < 0.9396). Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |- | ||
| + | |[[File:M9S_(3HB).png|thumbnail|center|450px|<b>Bdh2 with no pelB secretion tag growth.</b> Bdh2 MG1655 cells were grown in M9S media to gauge growth, the growth does not differ from the control as p = 0.5543. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |||
| + | |[[File:M9S_(3HB)_fluor.png|thumbnail|center|450px|<b>Bdh2 with no pelB secretion tag fluorescence.</b> MG1655 cells with bdh2 were grown in [https://2013.igem.org/Team:Imperial_College/Protocols#M9_minimal_and_supplemented_media M9S] and then tested for fluorescence by measuring their GFP output. There is significant fluorescence as p < 0.0001. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
|} | |} | ||
| - | |||
| - | < | + | <span style="color:#f00">'''Conclusion:'''</span> We can conclude that there is a slight increase in the growth rate of bdh2 containing cells at 10,000 uM 3HB. This suggests that bdh2 is able to function as expected and produces acetoacetate which is used by the cell's central metabolic pathways for growth. At 100uM or 100,000 uM, we did not observe any differences in growth. This could be be because the rate limiting step at low concentrations could be the uptake of 3HB rather than it's conversion to acetoacetate. We hope that we can observe an increase in growth if we add the putative permease we designed into the system. At elevated bdh2 concentration, there is a drop in the growth rate in both bdh2 and control cells, probably because of toxicity issues. In addition, fluorescence is highly elevated in bdh2 compared to empty vector control. |
| - | < | + | <h3> 3HB permease enhance cell growth in 3HB </h3> |
| - | + | In module 2, we make P3HB bioplastic from 3HB monomers. It is essential for the cells to take up a relatively high concentration of 3HB. Here we test the effect of having 3HB permease in M9 supplement media, where 3HB is the sole carbon source. | |
| - | + | ||
| - | + | ||
| - | |||
| - | < | + | [[File:Imperial_permease.jpg|thumbnail|center|600px|x|<b> 3HB sole carbon source growth in M9S with 3HB permease.</b> MG1655 with 3HB permease can grow better in M9 supplement media using 3HB as the sole carbon source, compared to MG1655 empty vector. 3HB concentrations were tested at 4 levels - 1 mM, 10 mM, 50 mM and 100 mM. Growth inhibitory effect exhibited at 100 mM of 3HB. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=3. Figure made by Imperial College London 2013 iGEM.<br><b><span style="color:#FF8000"><font size="6">Performed after European Jamboree</font size="6"></span></b>]] |
| - | + | <h3>Acetoacetate</h3> | |
| - | < | + | {| class="wikitable" style="margin: 1em auto 1em auto;" |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | < | + | |[[File:AA_sole_carbon_source.png|thumbnail|center|450px|<b>phaCAB in [https://2013.igem.org/Team:Imperial_College/Protocols#M9_minimal_and_supplemented_media M9 minimal and M9 supplemented media] with acetoacetate as sole carbon source.</b> Both media have a similar trend with increased acetoacetate concentration. Of interest is phaCAB in M9S at 10 mM, as this appears to be a local optimum, indeed this is seen in M9M grown phaCAB, while empty vector at 10 mM grows much less in comparison. An ANOVA for this data gives (<i>F</i> <sub>3,12</sub> = 1.669, p < 0.2262), which means that there is no significant difference between the data. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] |
| - | < | + | |
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| + | |[[File:AA_all.png|thumbnail|center|450px|<b>phaCAB promoter constructs in M9S with acetoacetate as sole carbon source.</b> Empty vector shows the least growth of all the constructs of MG1655. The best performing construct is hybrid phaCAB. According to ANOVA analysis, there is no statistical significance between the different phaCAB promoter constructs or EV (<i>F</i> <sub>3,12</sub> = 3.086, p < 0.068) with regard to growth on acetoacetate as a sole carbon source. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |} | ||
| + | <b>Conclusion: phaCAB grown in acetoacetate grows very well at 10 mM in M9S, this is confirmed with the new promoter constructs whereby all phaCAB growth much better than empty vector.</b> | ||
| + | |||
| + | <h3>Compiled sole carbon growth</h3> | ||
| + | <h4>phaCAB on M9 minimal and supplemented media</h4> | ||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| + | |||
| + | |[[File:M9M_phaCAB.png|thumbnail|center|450px|<b>phaCAB growth in sole carbon sources under M9M.</b> [https://2013.igem.org/Team:Imperial_College/Protocols#M9_minimal_and_supplemented_media M9 minimal] grown with phaCAB shows that 3HB is a poor carbon source at all concentrations tested. In addition, acetoacetate is poor as well at low concentrations, however at 10 mM acetoacetate, there is a spike in growth whereby it outperforms glucose. Glucose itself permits strong growth, especially as the concentration is increased from 22 mM to 167 mM. This said, ANOVA analysis showed that there was no significant difference between these values (<i>F</i> <sub>2,6</sub> = 2.745, p < 0.1424). Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |||
| + | |[[File:M9S_phaCAB.png|thumbnail|center|450px|<b>phaCAB growth in sole carbon sources under M9S.</b> M9 supplemented media grown with phaCAB displays similar results to those seen in M9M media. 3HB is once again the poorest carbon source, while acetoacetate has an optimum at 10 mM. Glucose performs very well in this situation as a sole carbon source. When submitted to ANOVA analysis, the sole carbon sources showed significant difference from each other (<i>F</i> <sub>2,6</sub> = 8.622, p < 0.0172). Testing the null hypothesis that there is no difference between sole carbon sources reveals the following. Applying a t-test shows that while 3HB and acetoacetate are not significantly different as carbon sources (p = 0.2341), both 3HB and glucose (p = 0.0256) and acetoacetate and glucose (p = 0.0488) act differently as sole carbon sources. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |} | ||
| + | |||
| + | <b>Conclusion: M9 Supplemented media is much more effective at getting the MG1655 phaCAB cells to grow than M9 minimal. Glucose was the best growth substrate, followed by acetoacetate and 3HB last.</b> | ||
| + | |||
| + | <h4>phaCAB with novel promoter</h4> | ||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| + | |||
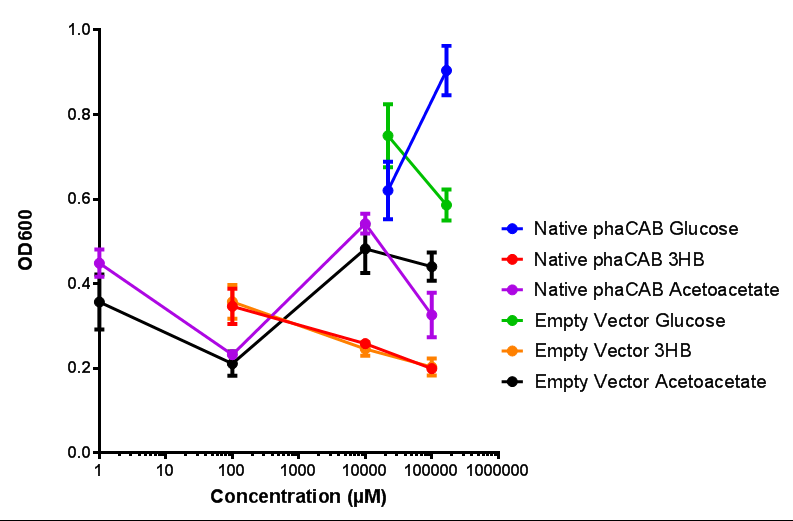
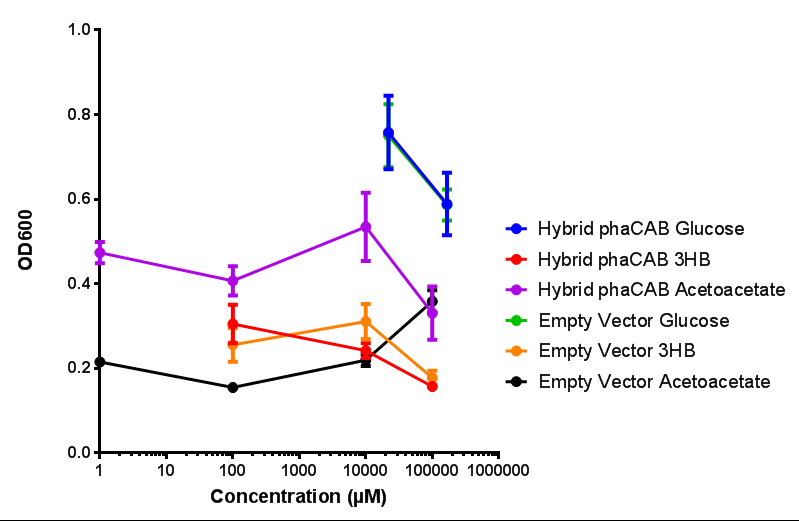
| + | |[[File:M9S_(native_phacab).png|thumbnail|center|450px|<b>Native promoter in phaCAB growth in sole carbon sources under M9S.</b> MG1655 <i>E. coli</i> transformed with native phaCAB compared to empty vector for sole carbon sources. The trends observed show that there is not much difference between EV and phaCAB for glucose (p = 0.6237), acetoacetate (p = 0.8781) and 3HB (p = 0.9901). ANOVA analysis concluded that null hypothesis, "The sole carbon sources tested have no effect on growth" must be rejected (<i>F</i> <sub>5,12</sub> = 6.938, p < 0.0029). This shows that there is a difference between individual carbon sources. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |||
| + | |||
| + | |[[File:M9S_(hybrid_phacab).png|thumbnail|center|450px|<b>Hybrid promoter in phaCAB growth in sole carbon sources under M9S.</b> MG1655 <i>E. coli</i> transformed with native phaCAB compared to empty vector for sole carbon sources. The trends observed show that there are differences between EV and phaCAB for glucose (p = 0.9715), acetoacetate (p = 0.0176) and 3HB (p = 0.8276). Indeed, hybrid phaCAB grows exceptionally well on acetoacetate, even at low concentrations compared to empty vector which only grows well at 100 mM 3HB. ANOVA analysis concluded that null hypothesis, "The sole carbon sources tested have no effect on growth" must be rejected (<i>F</i> <sub>5,12</sub> = 14.13, p < 0.0001). This shows that there is a difference between individual carbon sources. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |||
| + | |- | ||
| + | |||
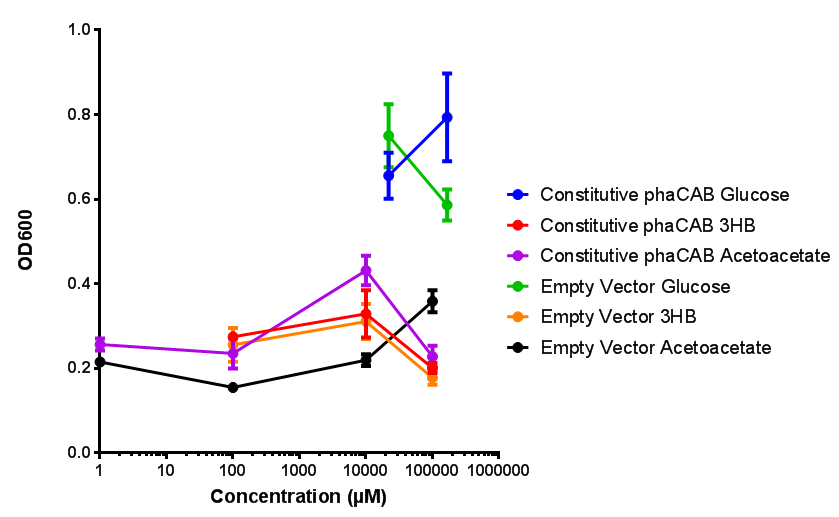
| + | |[[File:M9S(constitutive_phacab).png|thumbnail|center|450px|<b>Constitutive promoter in phaCAB growth in sole carbon sources under M9S.</b> MG1655 <i>E. coli</i> transformed with native phaCAB compared to empty vector for sole carbon sources. The trends observed show that there is not much difference between EV and phaCAB for glucose (p = 0.6511), acetoacetate (p = 0.4633) and 3HB (p = 0.7266). ANOVA analysis concluded that null hypothesis, "The sole carbon sources tested have no effect on growth" must be rejected (<i>F</i> <sub>5,12</sub> = 15.94, p < 0.0001). This shows that there is a difference between individual carbon sources. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM.]] | ||
| + | |} | ||
| + | <b>Conclusions: For native promoter and empty vector, there is no significant difference in growth for the same sole carbon source. There is a significant difference however between the different sole carbon sources used. A similar picture is observed with the constitutive promoter for phaCAB. By far, the most interesting result is seen with the hybrid phaCAB promoter. <span style="color:#f00">There is a very distinct difference between empty vector and the hybrid, whereby the hybrid phaCAB grows very well with acetoacetate up until 100 mM.</span></b> | ||
{{:Team:Imperial_College/Templates:footer}} | {{:Team:Imperial_College/Templates:footer}} | ||
Latest revision as of 03:18, 29 October 2013
Contents |
Growth and Toxicity Assays
This page includes all of our experimental growth, toxicity and sole carbon source assay data. OD600 in some experiments are larger than 1, as they were converted from absorbance at 600nm. Thus they are considered accurate.
Growth assays with different experimental media
In additional to standard LB and minimal media, several novel experimental media were developed in order to characterise Biobricks within a mixed waste/landfill setting. These media were characterised through an examination of pH and through an array of growth assays with the project chassis, E.coli (MG1655).
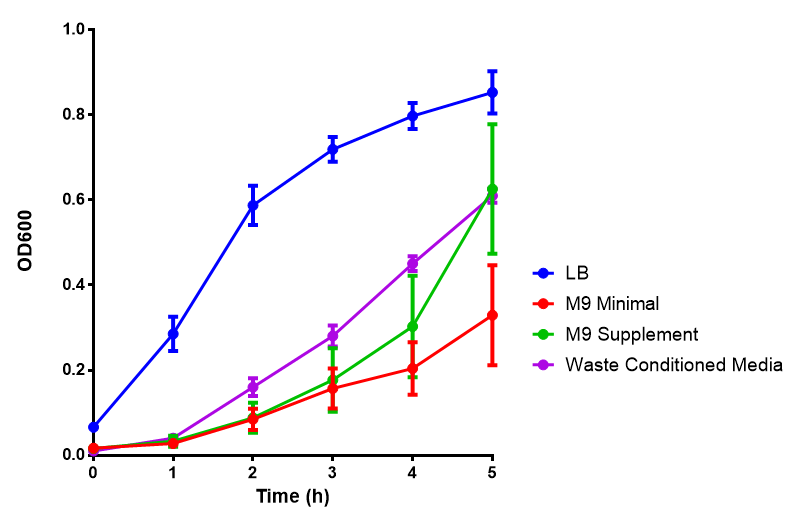 Media characterisation. E. coli strain MG1655 were transformed with a control plasmid and grown in different experimental media over a period of 5 hours. LB media, minimal media (M9M), supplemented minimal media (M9S), as described here or waste conditioned media (WCM), which is made from sterile filtrated mixed waste, see here. OD600 measured, error bars are S.E.M., n=4. Figure made by Imperial College London 2013 iGEM. |
Conclusion: MG1655 E. coli are viable and grow in all of our experimental medias. We have established a novel media that is optimised for characterisation of biobricks within a mixed waste/landfill context. LB as a long term media suffers from increased alkalinity over a period of a month
Long term waste growth assays
These assays were designed to test whether our chassis, E. coli (MG1655) could grow directly with waste over a long period of time.
Waste media
Conclusion: MG1655 E. coli are viable and grow on mixed waste alone. Therefore we have established that our chassis could survive in a mixed waste bio-reactor context, which is validation of our concept to industrially implement our system.
Waste conditioned media
These assays were designed to test whether our chassis, E. coli (MG1655) could grow with waste conditioned media (WCM) over a period of 24-48 hours. Waste conditioned media is a filter sterilised version of the waste media and was designed for several reasons; Firstly we were unsure whether mixed waste would be toxic to Ecoli and hence a less concentrated version may be more suitable and secondly large chunks of waste would prevent accurate OD600 measurements and therefore we decided to filter out the largest chunks.
 Photograph of waste conditioned media cultures mCherry stress biosensor (BBa_K639003) transformed MG1655 were grown with LB-WCM at 37ºC, with shaking for 48 hours. Figure made by Imperial College London 2013 iGEM. | |
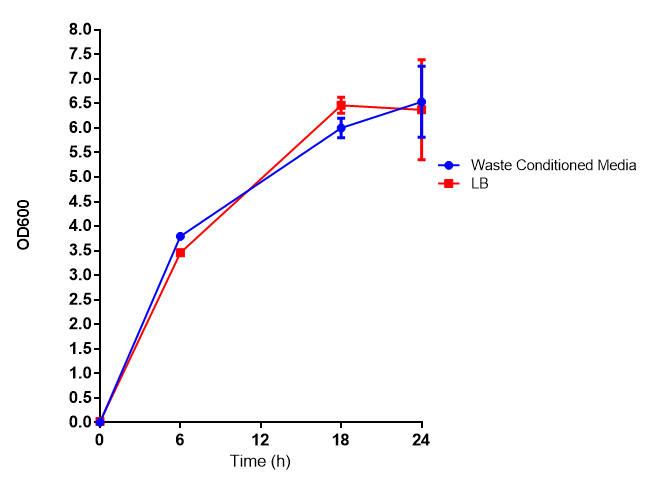 Growth assay in waste conditioned media (WCM). E. coli (MG1655) transformed with [http://parts.igem.org/Part:BBa_K639003 mCherry stress biosensor.] were grown with LB-based waste conditioned media at 37ºC, with shaking. Error bars represent S.E.M., n=4. Figure made by Imperial College London 2013 iGEM. | |
Conclusion: MG1655 transformed with either empty vector (EV) control or mCherry stress biosensor (BBa_K639003) vector are viable and can grow in waste conditioned media. Therefore waste conditioned media is an appropriate and novel experimental media with which to characterise biobricks within a mixed waste/landfill context. These data are also characterisation of an existing biobrick (BBa_K639003)
Growth and induction assays of our Biobricks in LB
Growth and induction assays of our project biobricks. Several of our constructs contain sfGFP within an operon and therefore fluorescence can be utilised to determine if expression is being induced by either addition of Arabinose or Xylose as appropriate to the construct. All of these characterisation assays were undertaken in LB media.
Empty Vector Control
Conclusions: Extracellular proteinase K fluoresces significantly more than uninduced extracellular proteinase K. None of the biobricks had significant decreases in their growth rates when induced by the respective inducer - be that Arabinose or Xylose. The remainder of the fluorescence assays did not show a significant difference between induction and no induction. This said, the data still requires normalising, this process might elucidate finally as to whether any other biobricks have significant fluorescence when induced. The empty vector control whether induced or not shows similar growth, proving its functionality as a control.
Growth assay characterisation of existing biobricks
Stress biosensor characterisation (BBa_K639003)
Originally we intended on using [http://parts.igem.org/Part:BBa_K639003 BBa_K639003] to detect whether our cells were stressed when grown with an array of potentially toxic plastics and degradation products. However, as the data below shows, the promoter is leaky and expresses mCherry in a non stressed state. As an alternative we utilised the stress sensor as a marker for our chassis E. coli (MG1655) for an array of qualitative and quantitative waste growth and toxicity assays.
Note: The stress sensor induces mCherry production through a mechanism involving the ppGpp stress response. Induction with IPTG bypassess this mechanism through an inhibition of LacI, resulting in mCherry expression.
Conclusion: The stress biosensor is activated when induced with IPTG but is leaky. In addition, growth decreases when cells are induced
phaCAB biobrick characterisation
LB
M9 Minimal
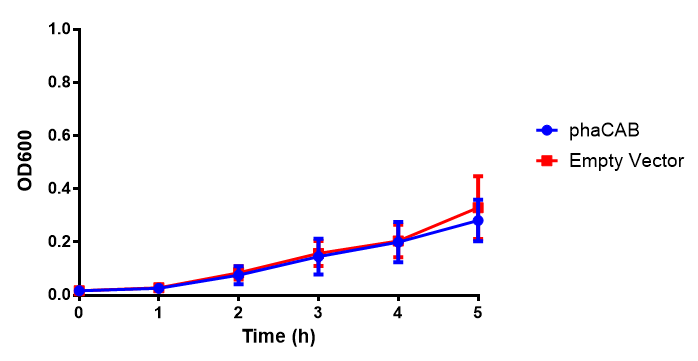
M9 Supplemented
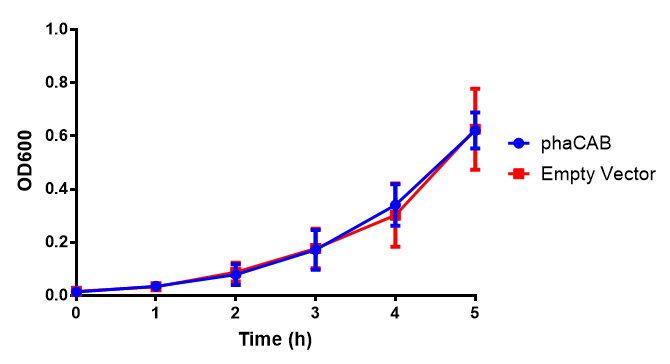
Conclusions: No difference between any of our phaCAB promoter constructs (native, hybrid, constitutive) when grown on LB compared to empty vector. In addition, there was no marked difference for native phaCAB in M9 minimal and M9 supplemented media, although the E. coli grew very well in supplemented idea.
pBAD characterisation
Characterisation of the strong pBAD promoter was a necessity to our project. We wanted to know whether or not it was inducing a significant increase in fluorescence of our constructs. We also wanted to know whether arabinose at the concentration we were inducing with (6 μM) was sufficient. Finally we wanted to look at whether or not glucose has an effect on growth. In order to address this we performed assays with pBAD in intracellular bdh2 in M9 minimal media. To tie it in with our project, we also used M9 minimal waste conditioned media (M9MWCM) to compare growth. Bdh2 is responsible for the synthesis of acetoacetate from 3HB and tagged with sfGFP.
The null hypothesis, glucose concentration does not influence growth of MG1655 in M9 minimal (M9M) media and M9 minimal waste conditioned media (M9MWCM) was tested using a two-tailed t-test. This must be rejected because p < 0.0001, thus glucose concentration has an influence on growth in M9M and M9MWCM. In addition to this, the null hypothesis: glucose concentration does not influence fluorescence in M9 minimal media must be rejected as p < 0.0001. Thus glucose concentration influences fluorescence.
Conclusions: Growth is significantly influenced glucose with regards to growth in M9M and M9MWCM. Bdh2 grown in glucose grow better in M9MWCM than M9M but are inhibited with increasing glucose concentration.
Glucose
Conclusions: Glucose has an optimum growth around 3-4%. In M9S, it does not give any growth advantage to cells cultured in higher glucose concentrations. Cells grown in M9S grew equally well as those grown in LB, possibly because of LB having a pH shift as it gets used up.
Plastic Toxicity Assays
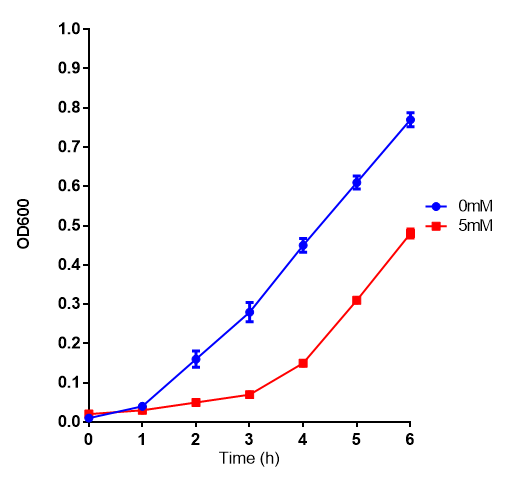
L-lactic Acid
Ethylene glycol
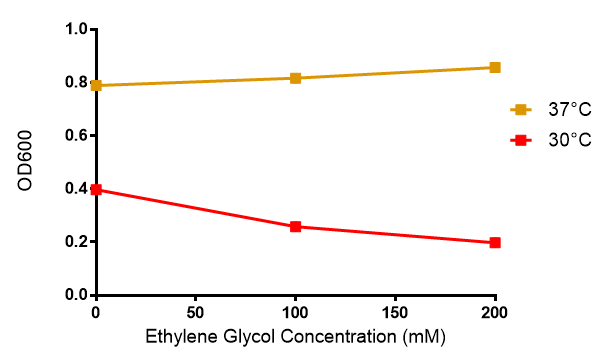
Reduced growth at 30°C likely due to decreased efficiency of MG1655 ethylene glycol break down enzymes. These enzymes (see UC Davis 2012) are endogenously expressed and detoxify Ethylene Glycol.
3-hydroxybutyrate (3HB)
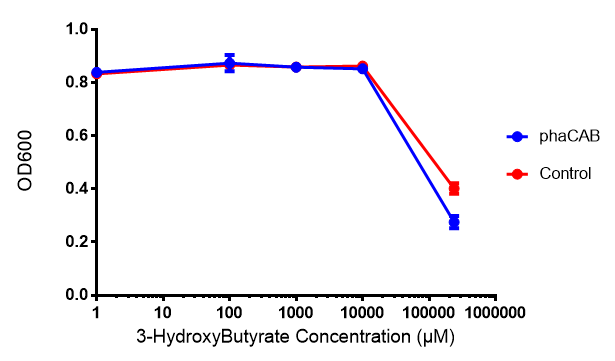
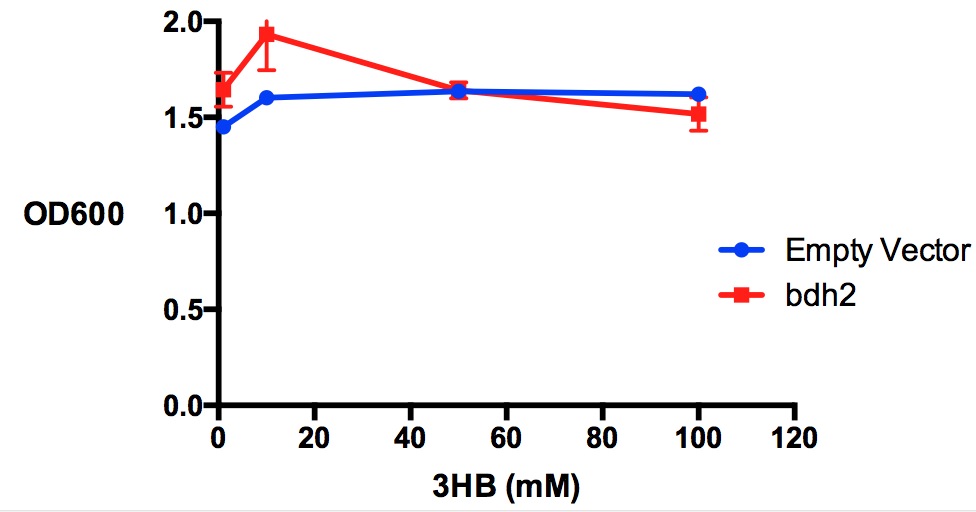
Performed after European Jamboree
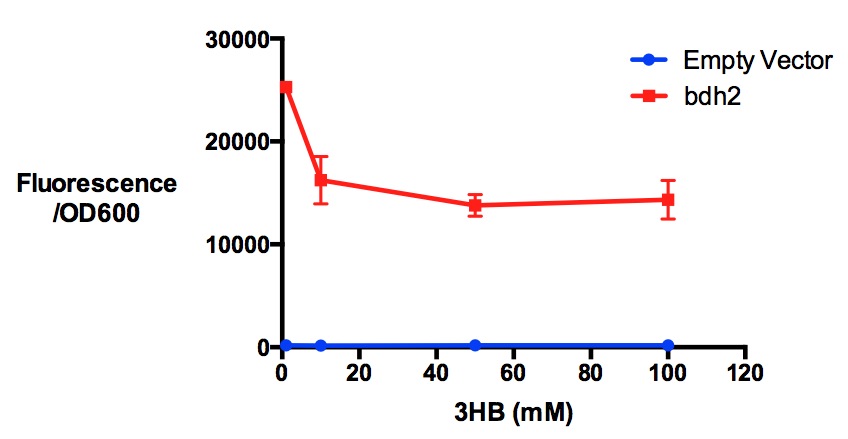
Performed after European Jamboree
Acetoacetate
Poly(3-hydroxybutyrate) P(3HB)
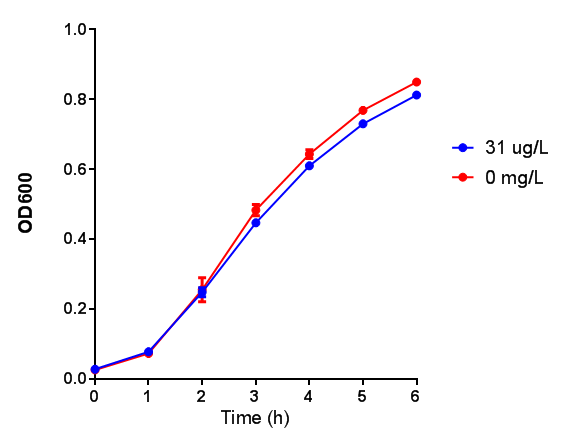
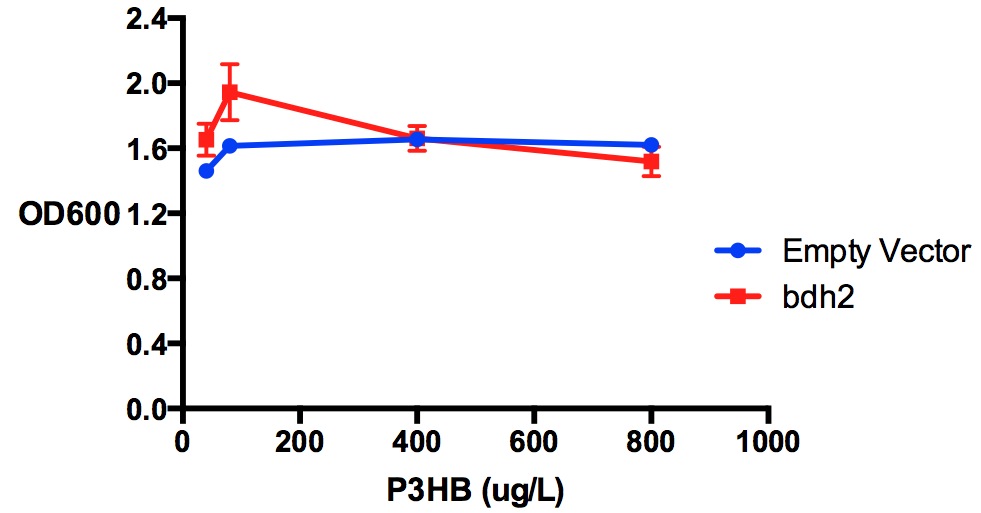
Performed after European Jamboree

Performed after European Jamboree
Poly(lactic acid) (PLA)
Conclusions: Elevated lactic acid results in a pronounced lag phase. At 37°C ethylene glycol does not have a significant effect on MG1655 growth. 3HB and acetoacetate are not toxic until their molarity is or exceeds 237 mM and 287 mM respectively. At the tested concentrations, neither PLA nor P(3HB) had a significant effect on growth.
Sole carbon source
bdh2 improves growth on 3HB
We have previously observed that MG1655 cells can survive in minimal media that contains 3HB as sole carbon source. This is evidence for that some uptake mechanism and a metabolic pathway is active at a low level in the cells. We hypothesised that the 3HB dehydrogenase could improve growth since it can convert 3HB into acetoacetate, a common metabolite in E.coli.
In order to test this, we run a growth experiment with various 3HB concentrations where the growth of cells containing bdh2 (BBa_K1149050) and Empty vector was recorded on a 96 well plate. We have calculated the growth rates from this data and plot it on the graph below.
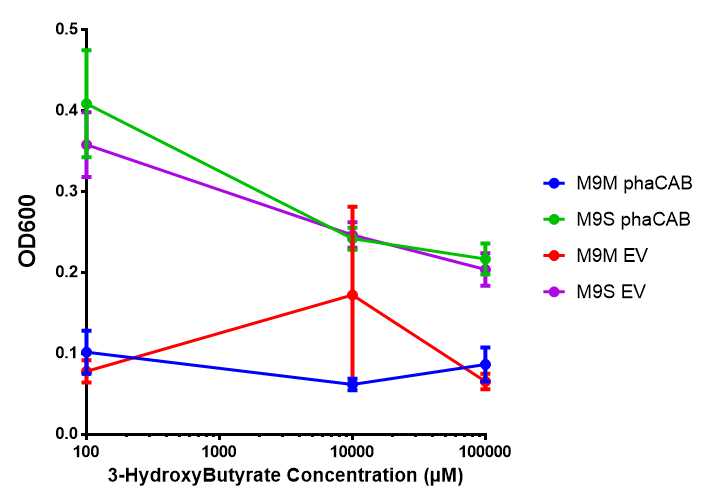 3HB sole carbon source growth in minimal media with phaCAB and EV. MG1655 phaCAB were grown in supplemented and minimal M9 media. Relative to the empty vector control, there was no significant difference in growth as t-test gave p = 0.8072 > 0.05. However, an ANOVA of the data gave (F 3,8 = 6.589, p < 0.0149), thus the null hypothesis, there is no difference between M9M and M9S must be rejected. Indeed if we look closer, we see that both M9M EV and M9S EV are significantly different from another (p = 0.045), as are M9M phaCAB and M9S phaCAB (p = 0.0284). Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM. | 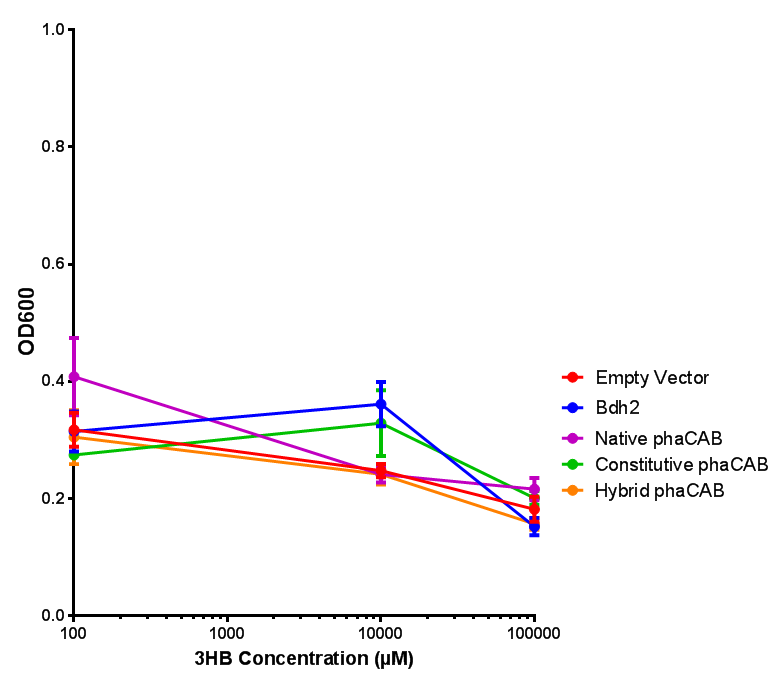 3HB sole carbon source growth in M9S with P3HB synthesis pathway genes. The 3 phaCAB promoter constructs are represented with native phaCAB, constitutive phaCAB and hybrid phaCAB. In addition bdh2 was tested to see whether its presence increased growth for the cells. This was completed with 3 3HB concentrations - 100 μM, 10 mM and 100 mM. ANOVA analysis shows that there is no significant difference between any of the constructs (F 4,10 = 0.1875, p < 0.9396). Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM. |
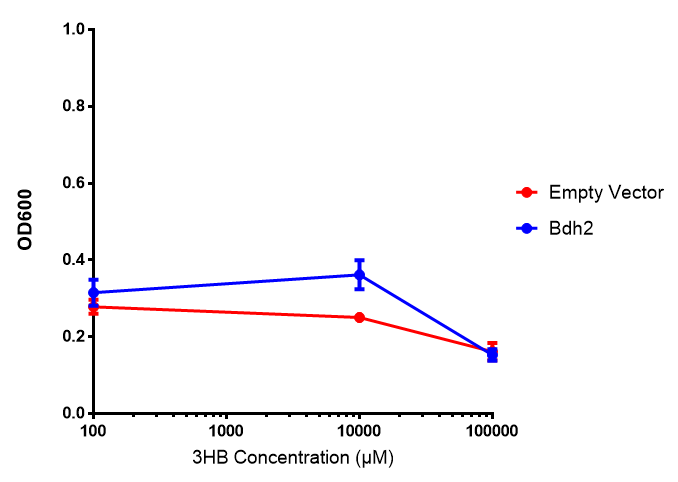 Bdh2 with no pelB secretion tag growth. Bdh2 MG1655 cells were grown in M9S media to gauge growth, the growth does not differ from the control as p = 0.5543. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM. | 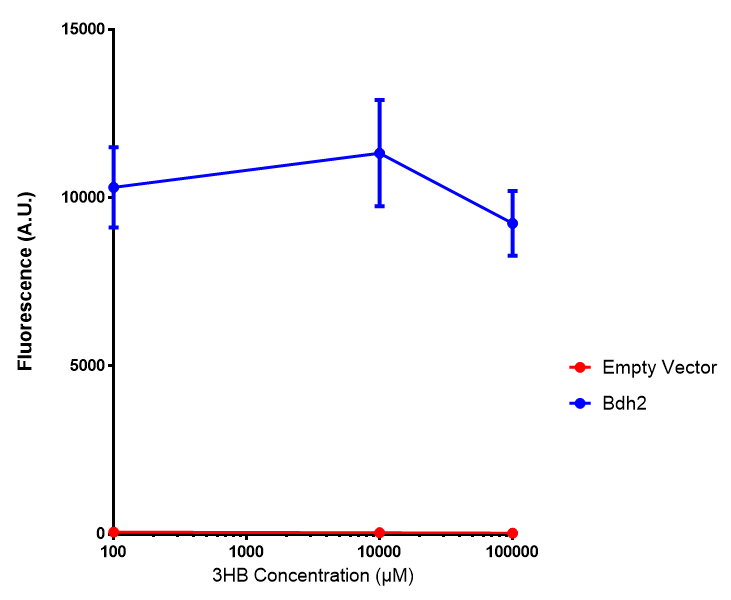 Bdh2 with no pelB secretion tag fluorescence. MG1655 cells with bdh2 were grown in M9S and then tested for fluorescence by measuring their GFP output. There is significant fluorescence as p < 0.0001. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM. |
Conclusion: We can conclude that there is a slight increase in the growth rate of bdh2 containing cells at 10,000 uM 3HB. This suggests that bdh2 is able to function as expected and produces acetoacetate which is used by the cell's central metabolic pathways for growth. At 100uM or 100,000 uM, we did not observe any differences in growth. This could be be because the rate limiting step at low concentrations could be the uptake of 3HB rather than it's conversion to acetoacetate. We hope that we can observe an increase in growth if we add the putative permease we designed into the system. At elevated bdh2 concentration, there is a drop in the growth rate in both bdh2 and control cells, probably because of toxicity issues. In addition, fluorescence is highly elevated in bdh2 compared to empty vector control.
3HB permease enhance cell growth in 3HB
In module 2, we make P3HB bioplastic from 3HB monomers. It is essential for the cells to take up a relatively high concentration of 3HB. Here we test the effect of having 3HB permease in M9 supplement media, where 3HB is the sole carbon source.

Performed after European Jamboree
Acetoacetate
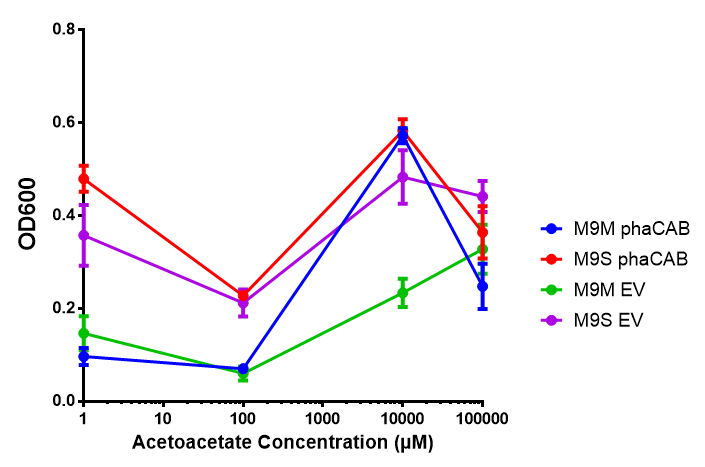 phaCAB in M9 minimal and M9 supplemented media with acetoacetate as sole carbon source. Both media have a similar trend with increased acetoacetate concentration. Of interest is phaCAB in M9S at 10 mM, as this appears to be a local optimum, indeed this is seen in M9M grown phaCAB, while empty vector at 10 mM grows much less in comparison. An ANOVA for this data gives (F 3,12 = 1.669, p < 0.2262), which means that there is no significant difference between the data. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM. | 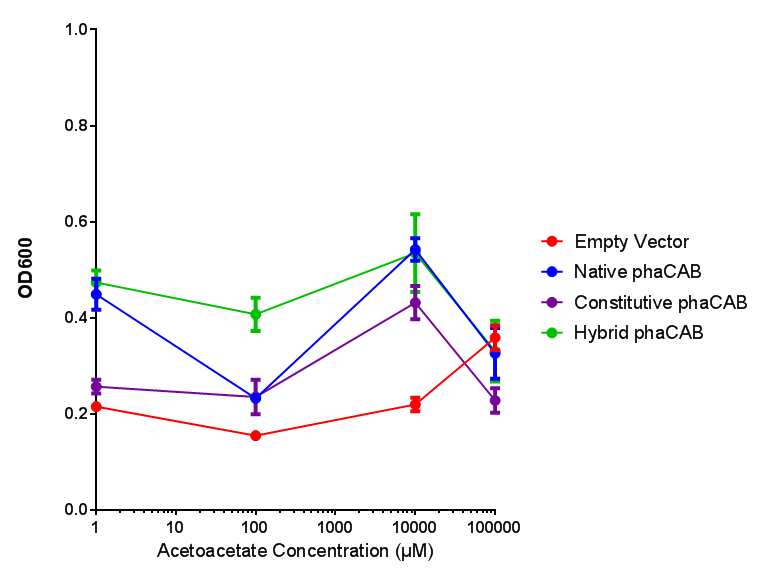 phaCAB promoter constructs in M9S with acetoacetate as sole carbon source. Empty vector shows the least growth of all the constructs of MG1655. The best performing construct is hybrid phaCAB. According to ANOVA analysis, there is no statistical significance between the different phaCAB promoter constructs or EV (F 3,12 = 3.086, p < 0.068) with regard to growth on acetoacetate as a sole carbon source. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM. |
Conclusion: phaCAB grown in acetoacetate grows very well at 10 mM in M9S, this is confirmed with the new promoter constructs whereby all phaCAB growth much better than empty vector.
Compiled sole carbon growth
phaCAB on M9 minimal and supplemented media
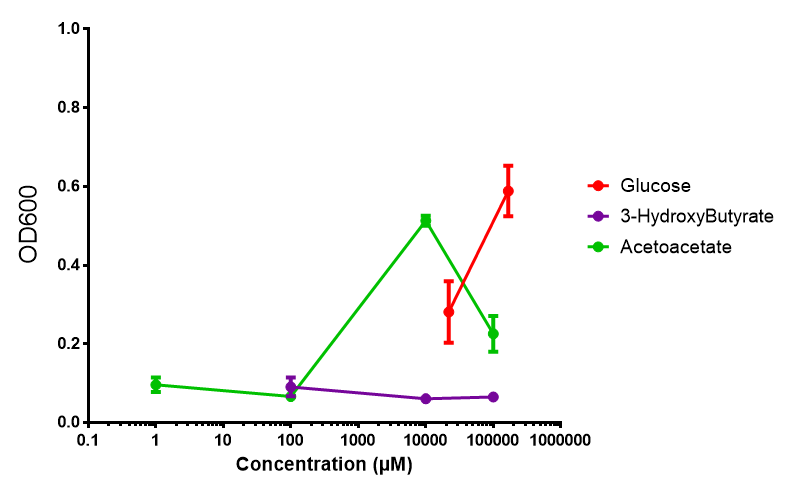 phaCAB growth in sole carbon sources under M9M. M9 minimal grown with phaCAB shows that 3HB is a poor carbon source at all concentrations tested. In addition, acetoacetate is poor as well at low concentrations, however at 10 mM acetoacetate, there is a spike in growth whereby it outperforms glucose. Glucose itself permits strong growth, especially as the concentration is increased from 22 mM to 167 mM. This said, ANOVA analysis showed that there was no significant difference between these values (F 2,6 = 2.745, p < 0.1424). Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM. | 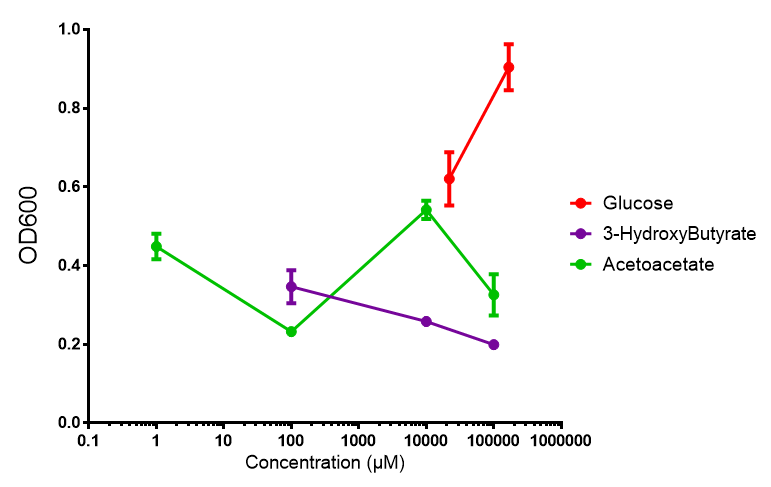 phaCAB growth in sole carbon sources under M9S. M9 supplemented media grown with phaCAB displays similar results to those seen in M9M media. 3HB is once again the poorest carbon source, while acetoacetate has an optimum at 10 mM. Glucose performs very well in this situation as a sole carbon source. When submitted to ANOVA analysis, the sole carbon sources showed significant difference from each other (F 2,6 = 8.622, p < 0.0172). Testing the null hypothesis that there is no difference between sole carbon sources reveals the following. Applying a t-test shows that while 3HB and acetoacetate are not significantly different as carbon sources (p = 0.2341), both 3HB and glucose (p = 0.0256) and acetoacetate and glucose (p = 0.0488) act differently as sole carbon sources. Data points show final time point after 6h growth for each concentration. Growth was at 37°C with shaking over 6h. Error bars are SEM, n=4. Figure made by Imperial College London 2013 iGEM. |
Conclusion: M9 Supplemented media is much more effective at getting the MG1655 phaCAB cells to grow than M9 minimal. Glucose was the best growth substrate, followed by acetoacetate and 3HB last.
phaCAB with novel promoter
Conclusions: For native promoter and empty vector, there is no significant difference in growth for the same sole carbon source. There is a significant difference however between the different sole carbon sources used. A similar picture is observed with the constitutive promoter for phaCAB. By far, the most interesting result is seen with the hybrid phaCAB promoter. There is a very distinct difference between empty vector and the hybrid, whereby the hybrid phaCAB grows very well with acetoacetate up until 100 mM.
 "
"