Team:Imperial College/PHB production
From 2013.igem.org
Margarita K (Talk | contribs) |
Margarita K (Talk | contribs) |
||
| (20 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Imperial_College/Templates:header}} | {{:Team:Imperial_College/Templates:header}} | ||
| + | __NOTOC__ | ||
<h1>PHB production</h1> | <h1>PHB production</h1> | ||
| - | |||
| - | |||
<h2 class="clear">Nile red staining</h2> | <h2 class="clear">Nile red staining</h2> | ||
| Line 9: | Line 8: | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
|[[File:EV_vs._phaCAB_red_-_reduced_background.PNG|thumbnail|right|400px|<b>Initial work with plastic synthesis in the native promoter.</b> Nile red staining was used to show expression of the plastic by fluorescence imaging. Control cells with empty vector are shown on the left, while native phaCAB transformed MG1655 is on the right.]] | |[[File:EV_vs._phaCAB_red_-_reduced_background.PNG|thumbnail|right|400px|<b>Initial work with plastic synthesis in the native promoter.</b> Nile red staining was used to show expression of the plastic by fluorescence imaging. Control cells with empty vector are shown on the left, while native phaCAB transformed MG1655 is on the right.]] | ||
| - | |[[File:27-9-13phaCABall.jpg|thumbnail|right|400px|<b> | + | |[[File:27-9-13phaCABall.jpg|thumbnail|right|400px|<b>phaCAB P(3HB) synthesis constructs transformed into MG1655</b> Strains were grown on Nile red plates, which stain the PHB strongly and fluoresce in presence of PHB. On the left are MG1655 cells with an empty vector (no fluorescence; no plastic), at the bottom is the native promoter (i.e. low fluorescence, some plastic). At the top and right we have our constitutive and hybrid promoter (respectively), which both show high expression and thus fluoresce very clearly.]] |
|} | |} | ||
<b>Conclusion: The red staining indicates the production of P(3HB). More importantly our new Biobricks [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149051 hybrid promoter phaCAB BBa_K1149051] and [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149052 constitutive phaCAB BBa_K1149052] produce more P(3HB) than the native phaCAB operon </b> | <b>Conclusion: The red staining indicates the production of P(3HB). More importantly our new Biobricks [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149051 hybrid promoter phaCAB BBa_K1149051] and [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149052 constitutive phaCAB BBa_K1149052] produce more P(3HB) than the native phaCAB operon </b> | ||
| + | |||
| + | <h2 class="clear">phaCAB OD-dry biomass calibration curve</h2> | ||
| + | |||
| + | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| + | |[[File:OD-Biomass.png|thumbnail|left|500px|<b>Optical Density(OD) of MG1655 containing phaCAB against dry biomass</b> ODs were measured for bacterial cultures before dessication. This plot was then used to calculate dry biomass from ODs in subsequent experiments. ]] | ||
| + | |} | ||
| Line 25: | Line 30: | ||
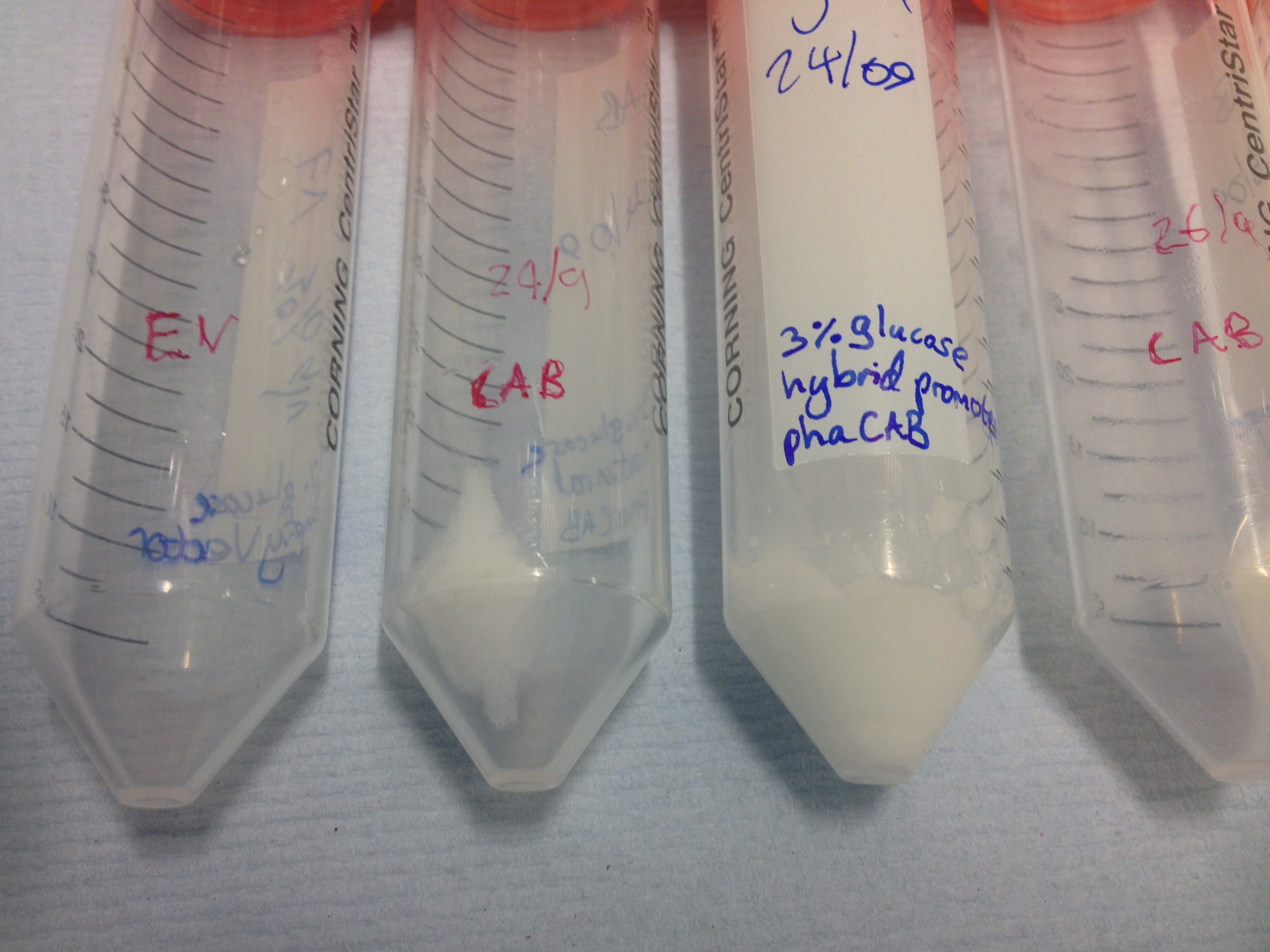
|[[File:Spun_down_hybrid_CAB_and_natural_CAB_3%25_glucose.jpg|thumbnail|right|450px|Cell pellets left after the centrifugation of 300ml LB media with 3% glucose. These pellets have slid back into the remaining supernatant.]] | |[[File:Spun_down_hybrid_CAB_and_natural_CAB_3%25_glucose.jpg|thumbnail|right|450px|Cell pellets left after the centrifugation of 300ml LB media with 3% glucose. These pellets have slid back into the remaining supernatant.]] | ||
|} | |} | ||
| + | |||
| + | <h2>Purification of P(3HB)</h2> | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| - | |[[File: | + | |[[File:EV-phaCAB-hybrid.JPG|thumbnail|left|500px|<b>Comparison of P3HB production </b> P3HB extracted from E.coli MG1655 with pSB1C3 containing, from left to right, nothing, natural phaCAB(BBa_K934001) and phaCAB expressed from the hybrid promoter, (BBa_K1149051). Each produced in 300ml cultures of LB with 3% glucose after one night growing at 37 degrees celsius.]] |
| - | + | ||
| - | + | ||
|} | |} | ||
{| class="wikitable" style="margin: 1em auto 1em auto;" | {| class="wikitable" style="margin: 1em auto 1em auto;" | ||
| - | |[[File: | + | |[[File:Moreplastic.JPG|thumbnail|left|500px|<b>Comparison of P3HB production </b> <b> (left)</b> 1.5ml tube, natural phaCAB (BBa_K934001) <b>(right)</b> 5ml tube, phaCAB expressed from the hybrid promoter, (BBa_K1149051).]] |
|} | |} | ||
| - | <h2 | + | <h2>P(3HB) from Mixed Waste</h2> |
| - | <p> | + | |
| + | <p class="clear">See [https://www.youtube.com/watch?v=4msf6EMNIrY here] how we convert trash to treasure, a longer more detailed version can be found [http://www.youtube.com/watch?v=1oXt_EfqmLU&feature=youtu.be here]</p> | ||
| + | |||
| + | |||
| + | [[File:Plasticfrommixedwaste.jpg|thumbnail|center|250px|P(3HB) extracted from phaCAB transformed MG1655 that were grown in M9M mixed waste.]] | ||
| + | |||
| - | <p> | + | <p> </p> |
| - | |||
{{:Team:Imperial_College/Templates:footer}} | {{:Team:Imperial_College/Templates:footer}} | ||
Latest revision as of 03:41, 29 October 2013
PHB production
Nile red staining
O/N cultures of MG1655 transformed with either control or phaCAB plasmid were spread onto LB-agar plates with 3% glucose and Nile red staining.
Conclusion: The red staining indicates the production of P(3HB). More importantly our new Biobricks [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149051 hybrid promoter phaCAB BBa_K1149051] and [http://parts.igem.org/wiki/index.php?title=Part:BBa_K1149052 constitutive phaCAB BBa_K1149052] produce more P(3HB) than the native phaCAB operon
phaCAB OD-dry biomass calibration curve
Extraction of P3HB
We extract P3HB using a technique which first disrupts the cell membranes and then degrades the remaining parts of the cell with bleach. See the protocols section for more details.
Once the cultures have been centrifuged and the supernatant poured off , the biomass is clearly seen as cell pellets.
Purification of P(3HB)
P(3HB) from Mixed Waste
See here how we convert trash to treasure, a longer more detailed version can be found [http://www.youtube.com/watch?v=1oXt_EfqmLU&feature=youtu.be here]
 "
"