Team:Yale/Project Bioassay
From 2013.igem.org
(Difference between revisions)
(→Aims for the Project) |
|||
| Line 22: | Line 22: | ||
=== Develop bioassay to screen PLA production === | === Develop bioassay to screen PLA production === | ||
| - | <center>[[File:Nile_red_PHB_stain.jpg|200px]]</center><br> | + | *We needed a way to detect the PLA once we produced it using the heterologous enzymes |
| + | **We decided to use the fluorescent dye Nile red, a intercellular lipid strain <br> | ||
| + | <center>[[File:Nile_red_PHB_stain.jpg|200px]]</center><br><br> | ||
| + | |||
| + | ====Positive Control==== | ||
| + | *We used the 2012 Tokyo Tech Biobrick (BBa_K934001) as a positive control to attempt to detect Nile red fluorescence on our plate reader (ex. 530nm, em. 590nm) | ||
| + | **This plasmid had the enzyme to synthesize P(3HB) a similar plastic PLA | ||
| + | <center>[[File:Tokyo Tech plasmid.jpg|400px]]</center><br><br> | ||
| + | |||
| + | ====Our Construct==== | ||
| + | *We then proceeded to test out plasmid in the plate reader. | ||
| + | **Cells were grown for 24 with both enzymes induced and in the presence of Nile red. The cells were washed and resuspended in PBS. The readings were normalized for optical density. | ||
| + | [[File:EcNR2 LB x20 wiki.jpg]] | ||
| + | |||
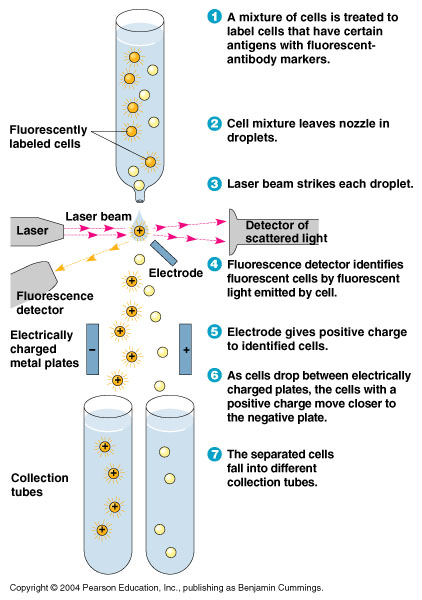
<center>[[File:FACS.jpg|400px]]</center> | <center>[[File:FACS.jpg|400px]]</center> | ||
Revision as of 04:00, 31 August 2013
| Project Overview | Validate PLA synthesis | Develop bioassay | Apply MAGE | Introduce export system | Make a bioplastic |
|---|
Contents |
Aims for the Project
- Engineer strains of E. coli to validate PLA synthesis
- Develop bioassay to screen PLA production
- Apply MAGE to optimize PLA production, guided by FBA
- Introduce type 1 secretion system to export and extract PLA
- Make a bioplastic
Develop bioassay to screen PLA production
- We needed a way to detect the PLA once we produced it using the heterologous enzymes
- We decided to use the fluorescent dye Nile red, a intercellular lipid strain
- We decided to use the fluorescent dye Nile red, a intercellular lipid strain

Positive Control
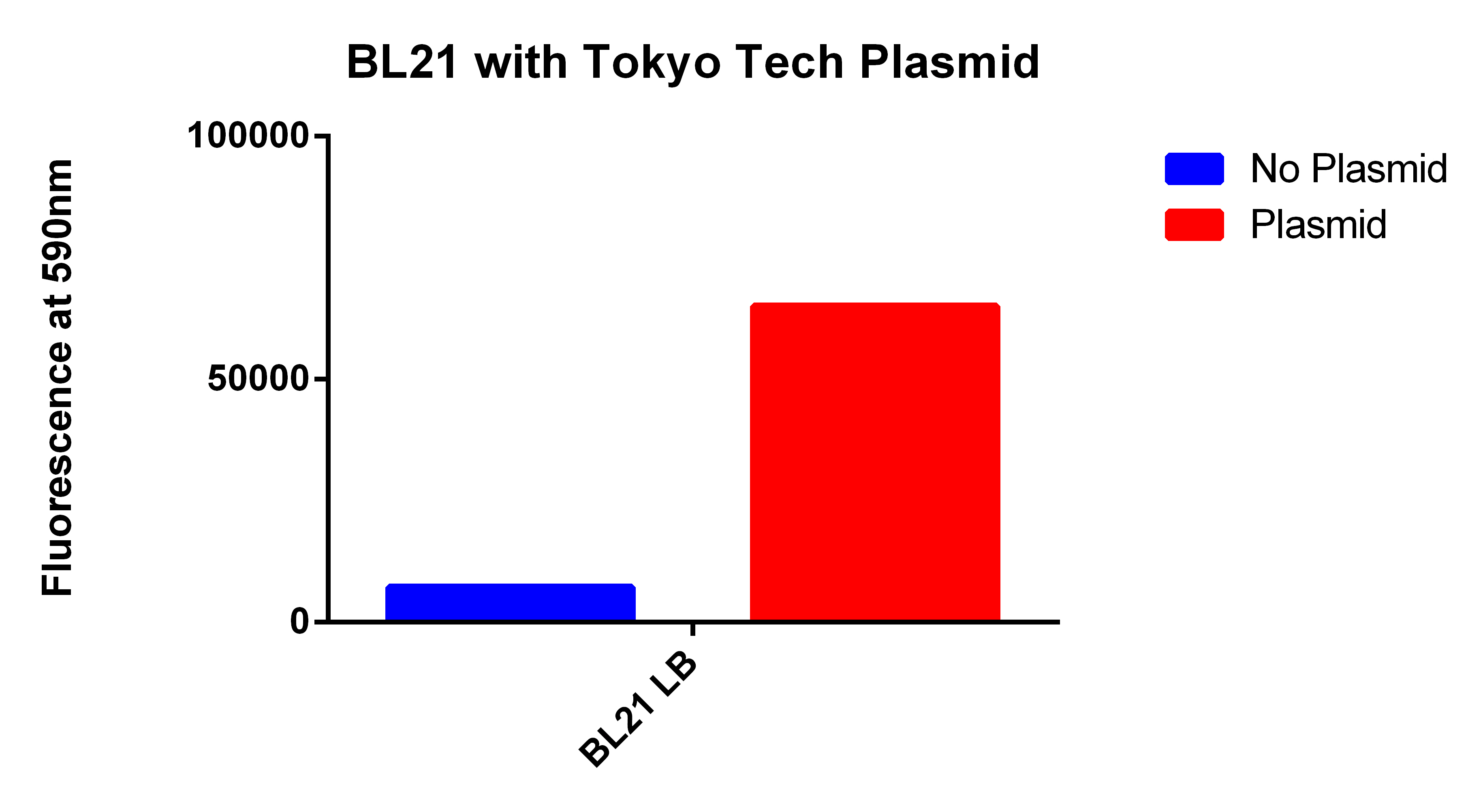
- We used the 2012 Tokyo Tech Biobrick (BBa_K934001) as a positive control to attempt to detect Nile red fluorescence on our plate reader (ex. 530nm, em. 590nm)
- This plasmid had the enzyme to synthesize P(3HB) a similar plastic PLA
Our Construct
- We then proceeded to test out plasmid in the plate reader.
- Cells were grown for 24 with both enzymes induced and in the presence of Nile red. The cells were washed and resuspended in PBS. The readings were normalized for optical density.
 "
"

