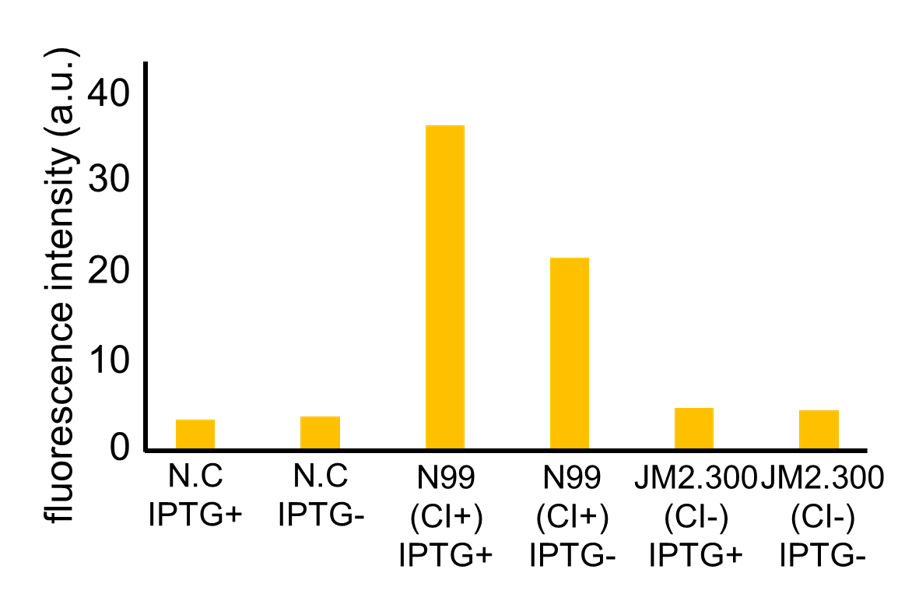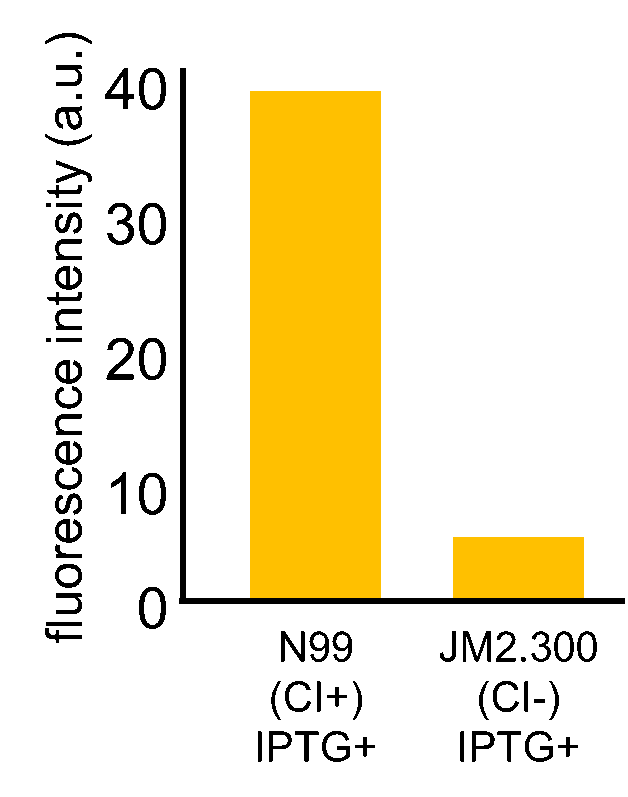Team:Tokyo Tech/Experiment/RM-lac Hybrid Promoter Assay
From 2013.igem.org
| (60 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{tokyotechmenudark}} | {{tokyotechmenudark}} | ||
| - | |||
| - | |||
<div id="text-area"><br> | <div id="text-area"><br> | ||
| + | <div class="box" id="title"> | ||
| + | <p style="line-height:0em; text-indent:0em;" name="top"><i>RM/lac</i> Hybrid Promoter Assay</p> | ||
| + | </div> | ||
<div class="box"> | <div class="box"> | ||
<h1>1. Introduction</h1> | <h1>1. Introduction</h1> | ||
<h2> | <h2> | ||
<p> | <p> | ||
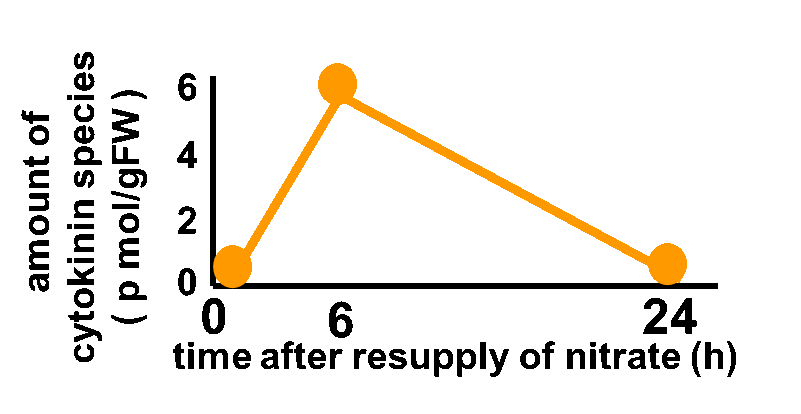
| - | In order to achieve the natural | + | In order to achieve the natural plants' temporal pattern for producing plant hormones in <i>E. coli</i>, we introduced an incoherent feed forward loop to our circuit, including a new hybrid promoter part ([http://parts.igem.org/Part:BBa_K1139150 BBa_K1139150]). Plants produce their hormones transiently rather than steadily (Takei et al., 2001) (Fig. 3-7-1). Moreover, continuous overexpression of hormones is harmful to plants (Thiman, 1937). Thus, we thought that it should be important to achieve this transient temporal pattern for producing plant hormones in <i>E. coli</i>, too. By applying the incoherent feed forward loop, the transient pulse of gene expression can be generated (Basu et al., 2003; Mangan et al., 2006). |
</p> | </p> | ||
| + | [[Image:Titech2013_rmlac_Fig_3-7-1.png|400px|thumb|center|Fig. 3-7-1. Temporal pattern for producing plant hormones in plants]] | ||
<p> | <p> | ||
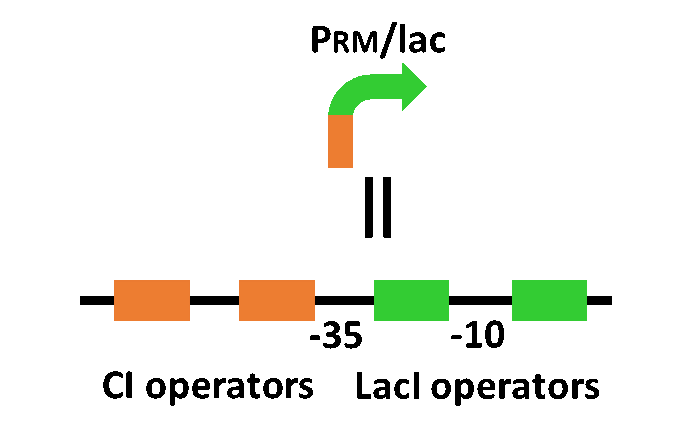
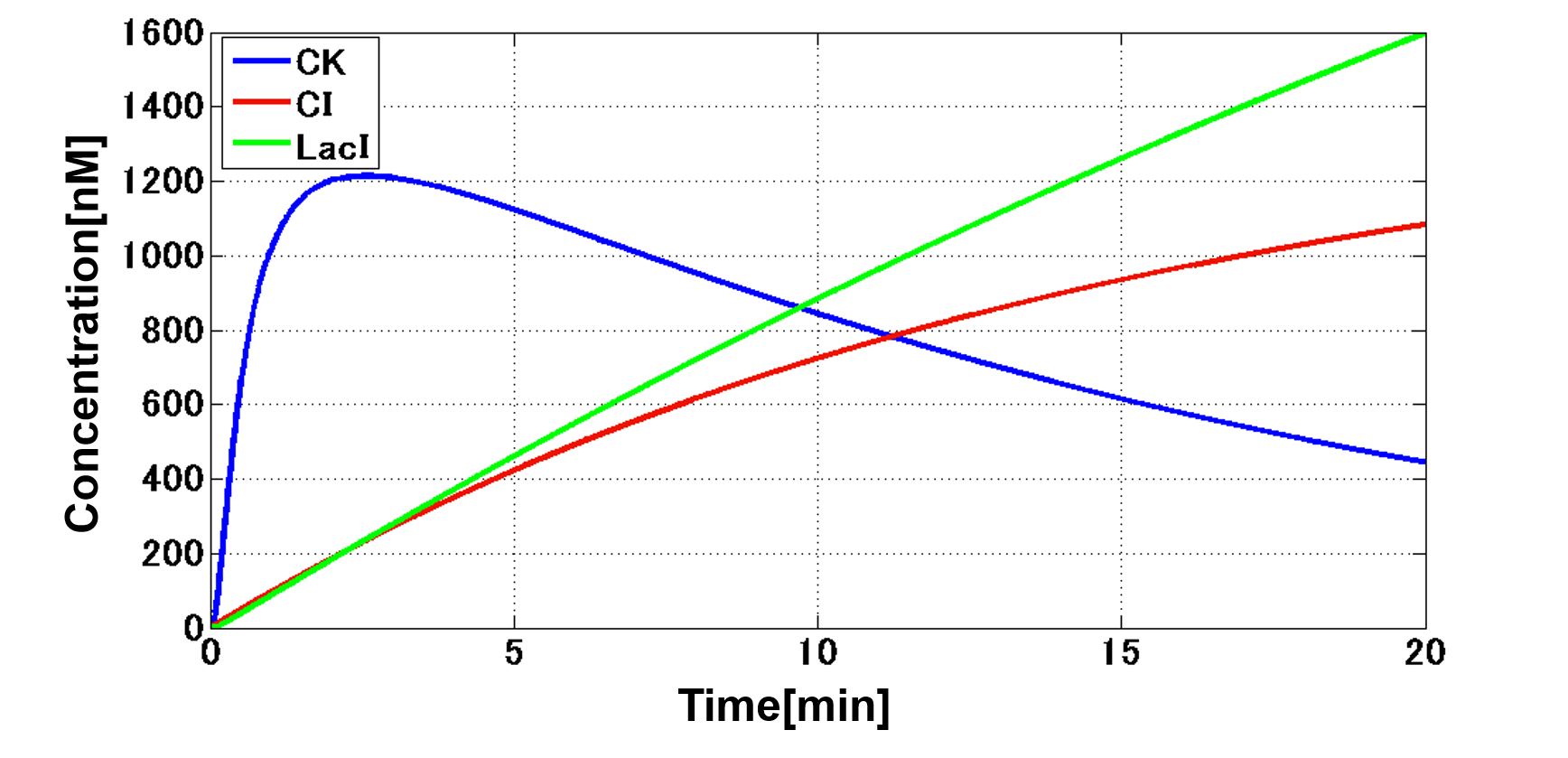
| - | + | Our designed system with an incoherent feed forward loop is shown in Fig. 3-7-2. We newly developed the <i>RM/lac</i> hybrid promoter, which is activated by CI and repressed by LacI (Fig. 3-7-3). We planned to ligate a hormone synthase part downstream of this hybrid promoter. Our mathematical model (Fig. 3-7-4) shows the pulse wave which the temporal pattern for plant hormone production should achieve (details about this model can be found [https://2013.igem.org/Team:Tokyo_Tech/Modeling/Incoherent_Feed_Forward_Loop#1._Introduction here]). While the <i>RM/lac</i> hybrid promoter activation by CI is a single-step reaction, the repression by LacI is a two-step reaction. Thus, the activation of the <i>RM/lac</i> hybrid promoter is faster than the repression. This time lag between the activation and the repression is important for generating a temporal pattern of plant hormone production. | |
</p> | </p> | ||
| + | <gallery widths="350px" heights="150px" style="margin-left:auto; margin-right:auto; text-align:center;"> | ||
| + | Image:Titech2013_rmlac_Fig_3-7-2.png|Fig. 3-7-2. Our designed system with an incoherent feed forward loop | ||
| + | Image:Titech2013_rmlac_Fig_3-7-3.png|Fig. 3-7-3. Our newly designed hybrid promoter | ||
| + | </gallery> | ||
| + | [[Image:Titech2013_rmlac_Fig_3-7-4.png|600px|thumb|center|Fig. 3-7-4. Our mathematical model for temporal pattern generation]] | ||
<p> | <p> | ||
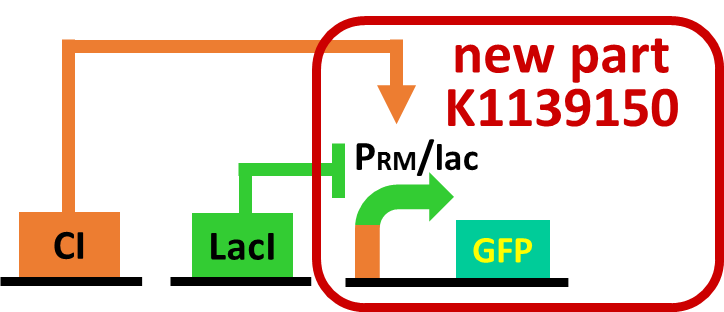
| - | As a first step to achieve this incoherent feed forward loop, we constructed | + | As a first step to achieve this system with an incoherent feed forward loop, we constructed a circuit shown in Fig. 3-7-5 to confirm that our new <i>RM/lac</i> hybrid promoter actually works. We set <i>GFP</i> as a reporter of the <i>RM/lac</i> hybrid promoter and introduced the part into <i>E. coli</i> . We verified that our <i>RM/lac</i> hybrid promoter was activated by CI and repressed by LacI with the following assay. |
</p> | </p> | ||
| + | [[Image:Titech2013_rmlac_Fig_3-7-5.png|400px|thumb|center|Fig. 3-7-5. The circuit we constructed including the new part]] | ||
</h2> | </h2> | ||
| - | + | <br> | |
| - | |||
<h1>2. Materials and Methods</h1> | <h1>2. Materials and Methods</h1> | ||
| + | <h2> | ||
| + | [[Image:Titech2013_rmlac_Fig_3-7-6.png|300px|thumb|right|Fig. 3-7-6. Constructions for our assay]] | ||
| + | </h2> | ||
<h3>2-1. Construction</h3> | <h3>2-1. Construction</h3> | ||
<h2> | <h2> | ||
| - | -pSB6A1-Ptet-GFP (N99)…positive control | + | <p>-pSB6A1-Ptet-<i>GFP</i> (N99)…positive control |
| - | < | + | </p> |
| - | < | + | <p>-pSB6A1-promoterless-<i>GFP</i> (N99)…negative control</p> |
| - | < | + | <p>-pSB6A1-PRM/lac-<i>GFP</i> (N99)…sample with CI* |
| - | < | + | </p> |
| + | <p>-pSB6A1--PRM/lac-<i>GFP</i> (JM2.300)…sample without CI | ||
| + | </p> | ||
| + | <p>* This N99 strain expresses CI from its genome constitutively. | ||
| + | </p> | ||
| + | |||
</h2> | </h2> | ||
<h3>2-2. Assay protocol</h3> | <h3>2-2. Assay protocol</h3> | ||
| + | <div style="margin-left:20px;"> | ||
<h2> | <h2> | ||
| - | + | <OL> | |
| - | < | + | <LI>Prepare overnight cultures of each cell at 37°C for 12 hours. |
| - | < | + | <LI>Take 30 µL of the overnight cultures into LB (3 mL) containing antibiotics (Amp 50 µg/mL) and 1 mM of IPTG* (→fresh culture) |
| - | < | + | <p>*We added IPTG in order to make sure to repress the expression of LacI derived from the <i>E. coli</i> genome. |
| + | </p> | ||
| + | <LI>After 4 hours of induction, measure the fluorescence intensity with a flow cytometer. | ||
| + | </OL> | ||
</h2> | </h2> | ||
| - | </div><br> | + | </div> |
| + | <br> | ||
| - | |||
<h1>3. Results</h1> | <h1>3. Results</h1> | ||
<h2> | <h2> | ||
<p> | <p> | ||
| - | Fig. 3- | + | Fig. 3-7-7 shows the fluorescence intensity detected by flow cytometer. Fig. 3-7-8 is the extracted data which shows the comparison of N99 (IPTG+, with constitutive CI) and JM2.300 (IPTG+, without CI). |
</p> | </p> | ||
| + | <gallery widths="350px" heights="300px" style="margin-left:auto; margin-right: auto; text-align:center"> | ||
| + | Image:Titech2013_rmlac_Fig_3-7-7.png|Fig. 3-7-7. Fluorescence intensity detected by flow cytometer | ||
| + | Image:Titech2013_rmlac_Fig_3-7-8.png|Fig. 3-7-8. Comparison of N99 and JM2.300 | ||
| + | </gallery> | ||
</h2> | </h2> | ||
| - | |||
| - | + | ||
<h1>4. Discussion</h1> | <h1>4. Discussion</h1> | ||
<h2> | <h2> | ||
<p> | <p> | ||
| - | N99 cells (CI+) showed higher fluorescence intensity than JM2.300 cells (CI-). From this | + | N99 cells (CI+) showed higher fluorescence intensity than that of JM2.300 cells (CI-). From this result, we assume that our <i>RM/lac</i> hybrid promoter was actually activated by CI. In addition, N99 (IPTG-) showed lower fluorescence than that of N99 (IPTG+). From this result, we can assume that our <i>RM/lac</i> hybrid promoter was repressed by LacI derived from the <i>E. coli</i> genome. |
</p> | </p> | ||
</h2> | </h2> | ||
| - | + | <br> | |
| - | + | <h1>5. References</h1> | |
| - | <h1>5. | + | |
<h2> | <h2> | ||
| - | Kentaro Takei, Hitoshi Sakakibara, Mitsutaka Taniguchi and Tatsuo Sugiyama. (2001) Nitrogen-Dependent Accumulation of Cytokinins in Root and the Translocation to Leaf: Implication of Cytokinin Species that Induces Gene Expression of Maize Response Regulator. Plant Cell Physiol. 42, 85–93 | + | <OL> |
| - | < | + | <LI>Kentaro Takei, Hitoshi Sakakibara, Mitsutaka Taniguchi and Tatsuo Sugiyama. (2001) Nitrogen-Dependent Accumulation of Cytokinins in Root and the Translocation to Leaf: Implication of Cytokinin Species that Induces Gene Expression of Maize Response Regulator. Plant Cell Physiol. 42, 85–93</LI> |
| - | Kenneth V. Thiman. (1937) | + | <LI>Kenneth V. Thiman. (1937) On The Nature of Inhibitions Caused by Auxin. American Journal of Botany. Vol. 24, No. 7, 407-412</LI> |
| - | < | + | <LI>Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. (2003) Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl. Acad. Sci. USA, 101, 6355-6360</LI> |
| - | Basu S, Mehreja R, Thiberge S, Chen MT, Weiss R. ( | + | <LI>S. Mangan, S. Itzkovitz, A. Zaslaver, U. Alon. (2006) The Incoherent Feed-forward Loop Accelerates the Response-time of the gal System of Escherichia coli . J. Mol. Biol. 356, 1073-1081</LI> |
| - | < | + | </OL> |
| - | S. Mangan, S. Itzkovitz, A. Zaslaver, U. Alon. (2006) The Incoherent Feed-forward Loop Accelerates the Response-time of the gal System of Escherichia coli . J. Mol. Biol. 356, 1073-1081 | + | |
| - | + | ||
</h2> | </h2> | ||
</div><br> | </div><br> | ||
| - | + | <html><div align="center"><a href="https://2013.igem.org/Team:Tokyo_Tech/Experiment/RM-lac_Hybrid_Promoter_Assay#top"><img src="https://static.igem.org/mediawiki/2013/f/f0/Titeh2013_backtotop.png" width="200px"></a></div></html> | |
| - | + | </div> | |
| - | </ | + | |
Latest revision as of 21:35, 28 October 2013
RM/lac Hybrid Promoter Assay
Contents |
1. Introduction
In order to achieve the natural plants' temporal pattern for producing plant hormones in E. coli, we introduced an incoherent feed forward loop to our circuit, including a new hybrid promoter part ([http://parts.igem.org/Part:BBa_K1139150 BBa_K1139150]). Plants produce their hormones transiently rather than steadily (Takei et al., 2001) (Fig. 3-7-1). Moreover, continuous overexpression of hormones is harmful to plants (Thiman, 1937). Thus, we thought that it should be important to achieve this transient temporal pattern for producing plant hormones in E. coli, too. By applying the incoherent feed forward loop, the transient pulse of gene expression can be generated (Basu et al., 2003; Mangan et al., 2006).
Our designed system with an incoherent feed forward loop is shown in Fig. 3-7-2. We newly developed the RM/lac hybrid promoter, which is activated by CI and repressed by LacI (Fig. 3-7-3). We planned to ligate a hormone synthase part downstream of this hybrid promoter. Our mathematical model (Fig. 3-7-4) shows the pulse wave which the temporal pattern for plant hormone production should achieve (details about this model can be found here). While the RM/lac hybrid promoter activation by CI is a single-step reaction, the repression by LacI is a two-step reaction. Thus, the activation of the RM/lac hybrid promoter is faster than the repression. This time lag between the activation and the repression is important for generating a temporal pattern of plant hormone production.
As a first step to achieve this system with an incoherent feed forward loop, we constructed a circuit shown in Fig. 3-7-5 to confirm that our new RM/lac hybrid promoter actually works. We set GFP as a reporter of the RM/lac hybrid promoter and introduced the part into E. coli . We verified that our RM/lac hybrid promoter was activated by CI and repressed by LacI with the following assay.
2. Materials and Methods
2-1. Construction
-pSB6A1-Ptet-GFP (N99)…positive control
-pSB6A1-promoterless-GFP (N99)…negative control
-pSB6A1-PRM/lac-GFP (N99)…sample with CI*
-pSB6A1--PRM/lac-GFP (JM2.300)…sample without CI
* This N99 strain expresses CI from its genome constitutively.
2-2. Assay protocol
- Prepare overnight cultures of each cell at 37°C for 12 hours.
- Take 30 µL of the overnight cultures into LB (3 mL) containing antibiotics (Amp 50 µg/mL) and 1 mM of IPTG* (→fresh culture)
*We added IPTG in order to make sure to repress the expression of LacI derived from the E. coli genome.
- After 4 hours of induction, measure the fluorescence intensity with a flow cytometer.
*We added IPTG in order to make sure to repress the expression of LacI derived from the E. coli genome.
3. Results
Fig. 3-7-7 shows the fluorescence intensity detected by flow cytometer. Fig. 3-7-8 is the extracted data which shows the comparison of N99 (IPTG+, with constitutive CI) and JM2.300 (IPTG+, without CI).
4. Discussion
N99 cells (CI+) showed higher fluorescence intensity than that of JM2.300 cells (CI-). From this result, we assume that our RM/lac hybrid promoter was actually activated by CI. In addition, N99 (IPTG-) showed lower fluorescence than that of N99 (IPTG+). From this result, we can assume that our RM/lac hybrid promoter was repressed by LacI derived from the E. coli genome.
 "
"